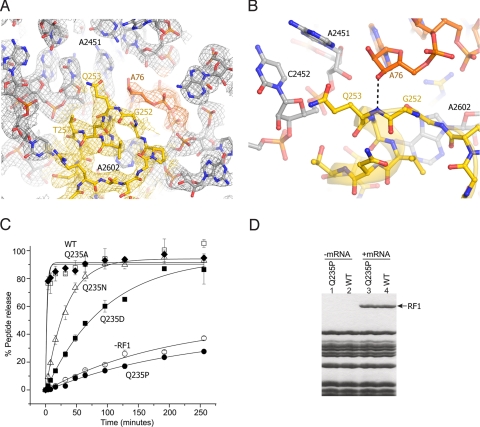
Fig. 4.
Interactions of the GGQ region of RF2 at the site of catalysis. (A) σA-weighted 3Fobs − 2Fcalc electron density for RF2 (yellow), P-site tRNA (orange) and 23S rRNA (gray), contoured at 1.7 σ. (B) Orientation of Gln-253 of the RF2 GGQ motif. The backbone amide nitrogen of Gln-253 is positioned to H-bond with the 3′-OH of A76 of P-site tRNA, whereas its side chain is oriented away from the reaction site. (C) The Q235P mutation abolishes peptide release activity of E. coli RF1. Model 70S termination complexes assembled with [35S]fMet-tRNAfMet in the P site and mRNA M0–27 (which contains an AUG codon followed a UAA stop codon), were incubated at 37 °C with (open squares) wild-type RF1, or the RF1 mutants (diamonds) Q235A (filled squares) Q235D (triangles) Q235N (filled circles) Q235P, or (open circles) no RF1. Peptide release was monitored by measuring the amount of [35S]fMet extracted into ethyl acetate at the indicated time points. Error bars indicate the range of values from 2 or 3 independent experiments, which were averaged and fit to single-exponential curves to determine the rates of peptide release. (D) Mutant Q235P RF1 binds normally in a stop-codon-dependent manner. Lanes 1 and 2, neither wild-type nor Q235P RF1 binds to ribosomes in the absence of mRNA. Lanes 3 and 4, in the presence of mRNA MO-27, both Q235P and wild-type RF1 bind stoichiometrically to ribosomes.