Fig. 3.
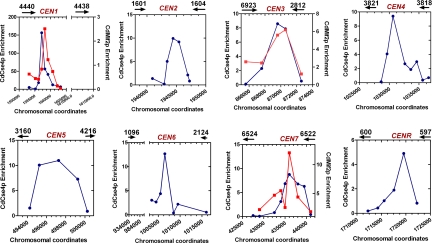
Two evolutionarily conserved key kinetochore proteins, CdCse4p (CENP-A homolog) and CdMif2p (CENP-C homolog) bind to the same regions of different C. dubliniensis chromosomes. Standard ChIP assays were performed on strains Cd36 and CDM1 (CdMif2-TAP-tagged strain) using anti-Ca/CdCse4p or anti-Protein A antibodies and analyzed with specific primers corresponding to putative centromere regions of C. dubliniensis to PCR amplify DNA fragments (150–300 bp) located at specific intervals as indicated (Table S2). Graphs show relative enrichment of CdCse4p (blue lines) and CdMif2p (red lines) that mark the boundaries of centromeric chromatin in various C. dubliniensis chromosomes. PCR was performed on total, immunoprecipitated (+Ab), and beads-only control (−Ab) ChIP DNA fractions (see Fig. S3 and Fig. S4). The coordinates of primer locations are based on the present version (May 16, 2007) of the Candida dubliniensis genome database. Enrichment values are calculated by determining the intensities of (+Ab) minus (−Ab) signals divided by the total DNA signals and are normalized to a value of 1 for the values obtained for a noncentromeric locus (CdLEU2) and plotted. The chromosomal coordinates are marked along the x-axis while the enrichment values are marked along the y-axis. Black arrows show the location of ORFs and arrowheads indicate the direction of transcription.