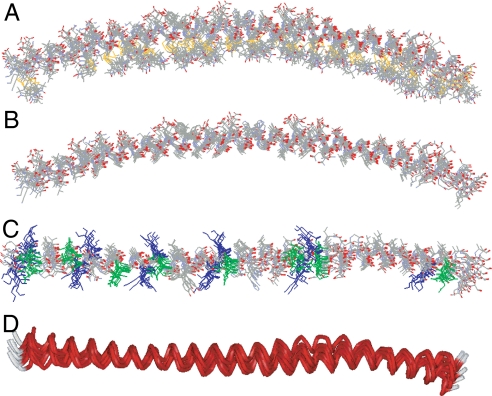
Fig. 3.
Refinement of the membrane-bound structure of α-synuclein by using SAMD with restraints from EPR data. (A) Overlay of nine structures obtained from SAMD calculations using the amino acids 9–89 fragment of α-synuclein, with spin labels added at 26 sites (Fig. S5). In the refined structures, the labels (identifiable by the S–S bond depicted in yellow) are generally positioned on the concave side of the curved protein structure (i.e., oriented into the membrane). The N terminus is on the left of the figure. (B) The refined labeled structures were converted to the respective α-synuclein structures by replacing each label with the normal amino acid in the α-synuclein sequence, with retention of the Cβ position in the label. (C) View of the overlaid structures from above the lipid surface. The 11 lysine residues (blue) are oriented approximately perpendicular to the helical axis, permitting potential interactions with the lipid phosphates. In contrast, the 8 glutamic acid residues (green) are oriented away from the membrane, on the top surface of the α-helix. (D) Ribbon view of the 9 overlaid structures (perspective similar to that in C), indicating the general goodness of fit of the structures and, most importantly, showing the subtle but reproducible superhelical twist in the α-helix that emerged in the SAMD calculations. We speculate that the superhelicity may allow the helix to adopt a 3-11 conformation that permits the helical axis to follow a curved surface over an extended length.