FIGURE 1.
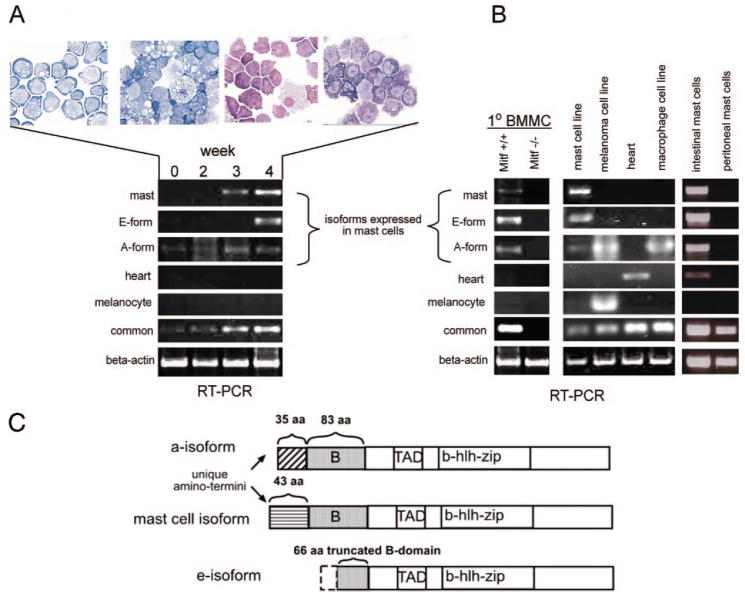
Mitf isoform expression in mast cells. A, RT-PCR analysis of in vitro-differentiated mast cells from murine embryonic stem cells shows expression of the mast cell isoform (mast), e-isoform (E-form), and a-isoform (A-form). Wright-Geimsa stain of cytospin preparations from differentiated cells at weekly time points above (original magnification, X60). Mast cell isoform and e-isoform expression is detectable with the appearance of morphologically identifiable mast cells at wk 3 and wk 4. B, RT-PCR analysis of specific Mitf isoforms from various tissue sources. Primary bone marrow-derived wild-type (Mitf+/+) mast cells (1°BMMC), intestinal mast cells, and the mast cell line, C57, show expression of mast cell, e- and a-isoforms. The heart isoform is also detected from intestinal cells. The melanoma cell line (B16), macrophage cell line (RAW), and heart tissue are used for controls for the heart, melanocyte, and a-isoforms. Specific isoforms are not detected from peritoneal mast cells, and no Mitf expression is detectable from Mitf−/− BMMCs. C, Schematic representation of protein domains of the Mitf isoforms expressed in mast cells show the unique amino-terminal domains of the mast cell isoform (▤, 43 aa) and the a-isoform (▨, 35 aa). The contiguous B-domain (
 , 83 aa) is shared between mast cell and a-isoform. The e-isoform harbors a truncated B domain amino terminus (66 aa). All isoforms share the transactivation domain (TAD), as well as the DNA binding basic domain and helix-loop-helix leucine zipper dimerization motif (b-hlh-zip).
, 83 aa) is shared between mast cell and a-isoform. The e-isoform harbors a truncated B domain amino terminus (66 aa). All isoforms share the transactivation domain (TAD), as well as the DNA binding basic domain and helix-loop-helix leucine zipper dimerization motif (b-hlh-zip).