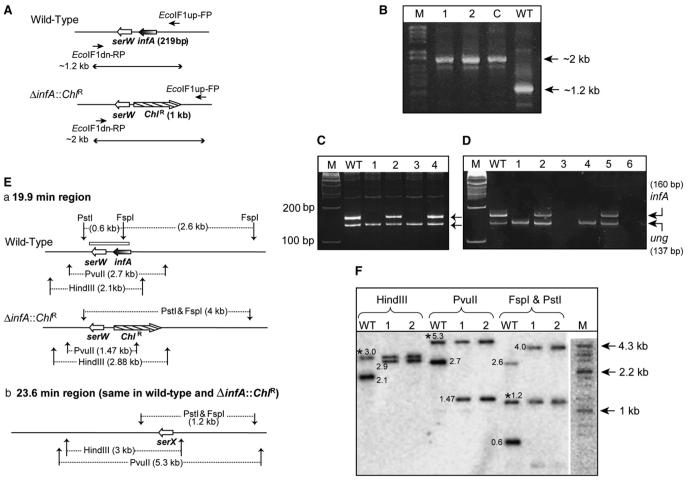
Figure 3. Generation and Analysis of E. coli ΔIF1 Strains.
(A) The organization of the infA locus. Lines with a single arrowhead indicate the location of primers (EcoIF1up-FP and EcoIF1 dn-RP) used, and lines with double arrowheads represent the expected length of amplicons with wild-type or ΔinfA::ChlR templates.
(B) Products of PCR with EcoIF1up-FP and EcoIF1 dn-RP; in lane WT, the template was from wild-type; in lane C from E. coli DY330 ΔIF1 + pTrc-MtuIF1; and in 1 and 2 from transductants obtained using E. coli TG1 ΔIF2 recipients harboring pACDH-IF2mt.
(C) PCR amplification of an internal fragment of infA open reading frame using IF1-RT-FP and IF1-RT-RP, and ung with EcoungSeq-FP and Ecoung-RT-RP. Lanes include the following: WT, wild-type; 1 and 3, ΔIF1ΔIF2; 2 and 4, ΔIF1ΔIF2 strains spiked with WT DNA. M denotes a 100 bp marker.
(D) RT-PCR to amplify an internal fragment of infA mRNA using IF1-RT-FP and IF1-RT-RP; and ung mRNA using EcoungSeq-FP and Ecoung-RT-RP. Lanes include the following: WT, wild-type; 1 and 4, ΔIF1ΔIF2 strains; 2 and 5, ΔIF1ΔIF2 spiked with RNA preparations from WT. Lanes 3 and 6 are reactions containing RNA from ΔIF1ΔIF2 strains without the reverse transcriptase. M denotes a 100 bp marker.
(E) (Ea) The 19.9 min region showing the infA locus and expected fragment lengths from digestion with various restriction enzymes. The box above infA/serW locus indicates the region corresponding to the probe used for Southern analysis. (Eb) The 23.6 min region of the E. coli genome containing serX, which remains unchanged in both strains but which was detected due to its identity to serW.
(F) Electronic autoradiography of the Southern blot using BioImager FLA5100 (Fuji Film, Japan). WT denotes the lane with wild-type genomic DNA, and 1 and 2 indicate lanes with genomic DNA of ΔinfA::ChlR ΔinfB (ΔIF1ΔIF2) transductants. M denotes the DNA size markers (λ HindII + HindIII). Bands common to the wild-type and the transductants due to hybridization to serX are indicated by an asterisk. Numbers next to the bands indicate their respective sizes in kilobases.