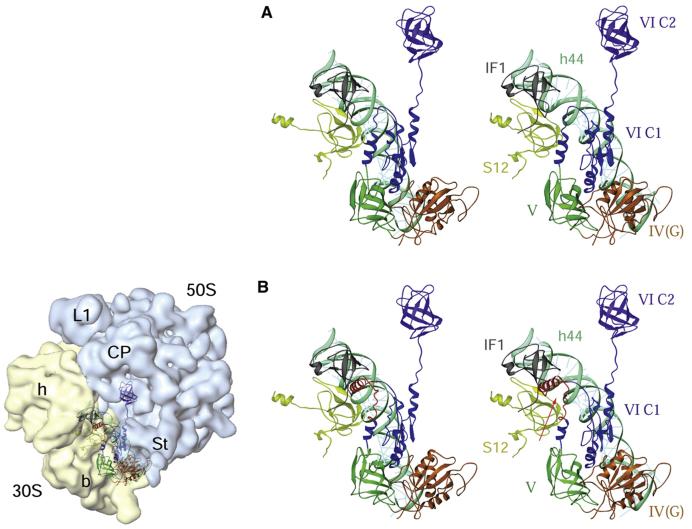
Figure 7. Interactions of Bacterial and Mammalian Mitochondrial IF2s with the E. coli 70S Ribosome.
(A) Stereo view presentation of the relative positions of the bacterial IF1 and IF2 on the E. coli 70S ribosome. The position of IF2 (identified by domains IV-VI, each depicted in different colors) was derived by docking the homology model of the factor into the cryo-EM map of an initiation complex (Allen et al., 2005), whereas the position of IF1 (gray) was obtained by docking the X-ray crystallographic structure of a 30S subunit•IF1 complex (Carter et al., 2001) into the cryo-EM map. Two 30S subunit components, protein S12 (greenish yellow) and 16S rRNA helix 44 (cyan), are also shown.
(B) A homology model of bovine IF2mt, aligned to the matching position of IF2 in (A), shows that the 37 amino acid insertion domain (red, indicated by an arrow) in the IF2mt overlaps partially with the position of IF1 in the bacterial ribosome, precluding the simultaneous binding of the two factors to the ribosome. The panel on the left of (B) depicts the orientation of the 70S ribosome, with 30S (semitransparent yellow) and 50S (semitransparent blue) ribosomal subunits and their prominent structural features identified: h and b mark the head and body, respectively, of the 30 subunit, while L1, CP, and St mark the protein L1, central protuberance, and protein L7/L12 stalk regions, respectively, of the 50S subunit (for details, refer to the Supplemental Data). Domains of IF2 are numbered according to Spencer and Spremulli (2005).