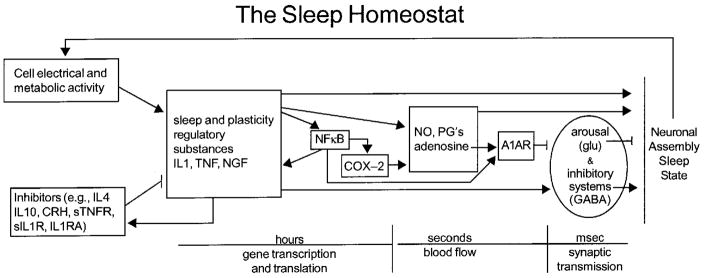
Figure 1. Molecular networks comprise the sleep homeostat.
Sleep regulatory substances (SRSs), including TNF and IL1 and nerve growth factor (NGF), are produced and released in response to neuronal activity and metabolism during wakefulness (see text). SRSs levels are influenced by positive and negative feedback including transcription factors such as NFκB, and cytokines and hormones. Cytokine production and actions involve transcription and translation events and occur over periods of hours or more. As such they offer a mechanism by which the CNS can track sleep/wake history. Their direct somnogenic actions involve labile substances with half lives of seconds such as NO and adenosine. Cytokines and their effector molecules in turn influence excitatory and inhibitor neurotransmitter systems to orchestrate sleep. The sleep homeostat possesses many of the characteristics of biological networks and engineered systems [reviewed 78,79]. It is modular in that several proteins are working in “overlapping co-regulated groups”. The molecular network is robust in that removal of any one of the components does not result in complete sleep loss. The network operates as a recurring circuit element in the sense that multiple molecular networks work in parallel, e.g. each within a different semiautonomous cortical column. Abbreviations, see text and IL-1 RA, IL1 receptor antagonist; sIL1R, soluble IL1 receptor; anti-IL1, anti-IL1 antibodies; CRH, corticotrophin releasing hormone; PGD2, prostaglandin D2; sTNFR, soluble TNF receptor; A1AR, adenosine A1 receptor; COX-2, cyclooxygenase 2; glu, glutamine acid; GABA, gamma amino buteric acid.