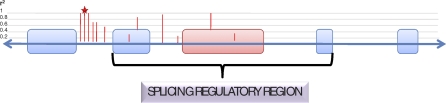
Figure 4. Methodological Details Evaluating the Proximity of a Detected sQTL and Its Region of LD to a Splicing Regulatory Region.
Red box represents an exon whose expression is correlated with the SNP indicated by the starred red bar. All SNPs in LD with this sQTL are shown by red bars and the height of the bar indicates the level of correlation (r 2) with the starred SNP. We assessed how often the range of LD for a given sQTL (defined by r 2 > 0.2 with the sQTL) extended into or surpassed the splicing regulatory region. This analysis was performed by evaluating all mRNA transcripts containing the exon regulated by the sQTL. The splicing regulatory region was defined as the genomic region from the start of the exon located upstream of the associated exon through the stop site of the downstream exon interrogated. If the exon was part of multiple transcripts the region including the most distal and proximal neighboring exons was defined the splicing regulatory region. If the affected exon was located at the beginning or end of the transcript then the range was truncated at the start or stop of the transcript, respectively. Finally, if a single SNP associated with more than one exon in a single transcript they were considered as a single entry in this analysis (i.e., sQTL LD needed only come in close proximity of one of the affected exons to be counted as a positive entry).