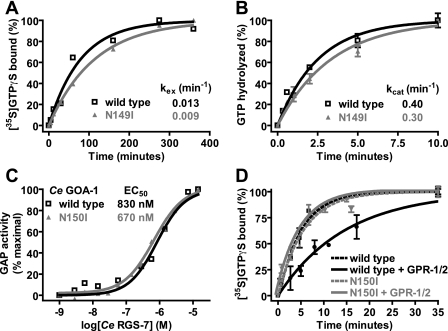
FIGURE 4.
Biochemical properties of GoLoco-insensitive Gα subunits. A, spontaneous nucleotide exchange rates (kex) of wild type (black) and N149I (gray)Gαi1·GDP were measured. A time course of specific binding of 100 nm Gα subunit to 1 μm GTPγS was determined using an [35S]GTPγS filter-binding assay. Data were fit to single exponential functions with rate constants as follows: wild type Gαi1 0.013 ± 0.002 min-1 and N149I Gαi1 0.009 ± 0.0004 min-1. B, spontaneous GTP hydrolysis rates (kcat) of wild type (black) and N149I (gray)Gαi1 were measured using [γ-32P]GTP hydrolysis assays. A time course of 32Pi (inorganic phosphate) production was determined using activated charcoal filtration. Data were fit to single exponential functions with rate constants as follows: wild type Gαi1 0.40 ± 0.003 min-1 and N149I Gαi1 0.30 ± 0.003 min-1. C, GTPase-accelerating protein (GAP) activity of C. elegans RGS7 on 200 nm wild type and N150I-mutated C. elegans GOA-1 was measured using 100 nm BODIPYFL-GTP and fluorescence spectroscopy. Data were fit to the four parameter logistic equation to determine EC50 values of RGS-7 GAP activity (95% confidence intervals in parentheses) as follows: wild type GOA-1, 830 (570-1300) nm; N149I GOA-1, 670 (580-790) nm. D, GDI effect of the C. elegans GPR-1/2 GoLoco motif on C. elegans GOA-1 was quantified using [35S]GTPγS filter binding. Time courses were obtained by preincubating 100 nm GOA-1 (wild type or N150I) with either buffer or 10 μm GPR-1/2 GoLoco motif peptide for 5 min. Samples were then added to 1 μm GTPγS, and specific [35S]GTPγS binding was quantified by filtration and scintillation counting. Data were fit to exponential association functions (95% confidence intervals in parentheses) as follows: wild type GOA-1 alone, 0.202 (0.160-0.240) min-1; wild type GOA-1 + GoLoco peptide, 0.068 (0.055-0.081) min-1; N150I GOA-1 alone, 0.178 (0.150-0.210) min-1; N150I GOA-1 + GoLoco peptide, 0.194 (0.140-0.250) min-1.