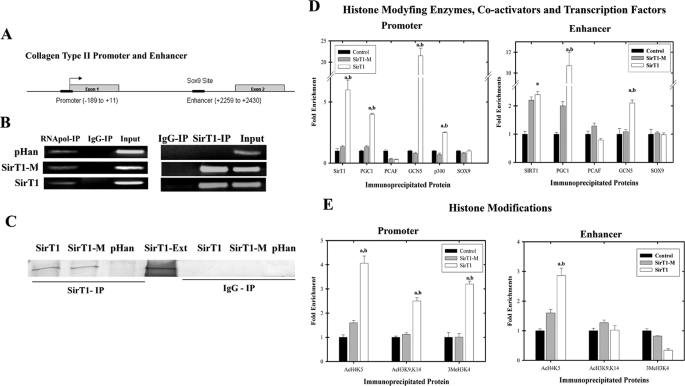
FIGURE 4.
Enzymatically active SirT1 recruits GCN5, PGC1α, and p300 to the collagen 2(α1) promoter and elevates histone modifications at this site. A, the collagen 2(α1) promoter and enhancer positions used in ChIP assays are indicated (black bars). B, chondrocytes stably expressing SirT1, SirT1-M, and the pHan control were processed for ChIP analyses using antibodies for SirT1, GAPDH, and an IgG control. PCR was then performed with primers flanking the enhancer within the collagen 2(α1) first intron (SirT1 IP). In the right panel, as an additional control, RNA polymerase II was immunoprecipitated, and the GAPDH promoter was amplified. The RT-PCR products shown are representative of three separate experiments (n = 3). C, protein extracts generated from the ChIP analyses were immunoblotted with SirT1. SirT1-Ext indicates protein extracts isolated from SirT1 stably expressing cells. D and E, ChIP analyses as in B using primers for either the enhancer or promoter sequence of collagen 2(α1) were carried out with the indicated cell lines. The chromatin was immunoprecipitated with antibodies specific for SirT1, PGC1α, GCN5, PCAF, p300, and Sox9 (D) or antibodies specific for AcH3K9/K14, AcH4K5, and 3MeH3K4 (E). The DNA products were amplified by quantitative PCR. An IgG control for D and E showed no PCR product and was subtracted from the equivalent antibody ChIP value. The error bars in the graphs indicate the S.D., and the statistical significance is indicated by an a (SirT1 compared with pHan) or b (SirT1-M compared with SirT1-M) (LSD, p < 0.05). The displayed immunoblots are representative of four experiments. qPCR was performed in triplicates using IPs from 2 distinct cell lines.