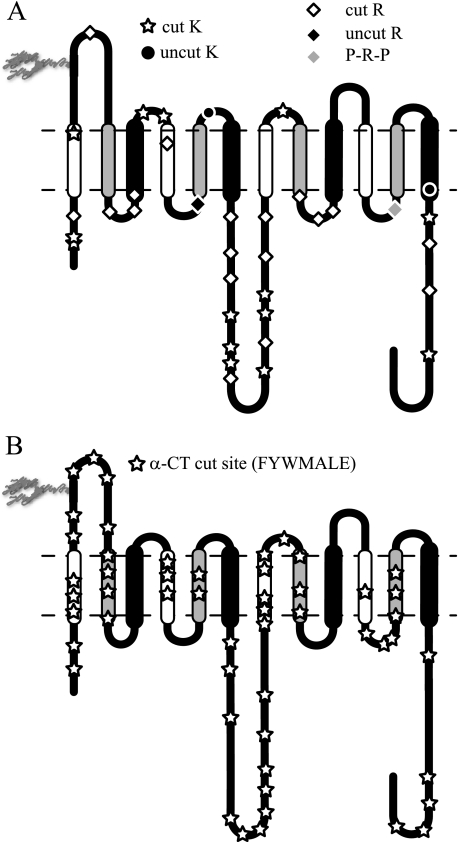
FIGURE 2.
Topography of protease-accessible sites. Membrane-resident GLUT1 was
digested with trypsin or α-chymotrypsin and then analyzed by reverse
phase HPLC-ESI-MS/MS. Peptides containing the indicated cleavage sites were
positively identified by MS/MS. A, GLUT1 contains 35 potential
trypsin cleavage sites (16 lysine residues and 19 arginine residues).
Thirty-two are cleaved by trypsin.
 , observed lysine cleavage
sites; •, lysine residues not observed as cleavage sites; ⋄,
observed arginine cleavage sites; ♦, arginine residues not observed as
cleavage sites. Arg400 (gray ♦) is flanked by N- and
C-terminal proline residues and is not a potential trypsin cleavage site.
B, GLUT1 contains 197 potential α-chymotrypsin cleavage sites
(Phe, Tyr, Trp, Leu, Met, Ala, and Glu). The 52 detected cleavage sites are
indicated by
, observed lysine cleavage
sites; •, lysine residues not observed as cleavage sites; ⋄,
observed arginine cleavage sites; ♦, arginine residues not observed as
cleavage sites. Arg400 (gray ♦) is flanked by N- and
C-terminal proline residues and is not a potential trypsin cleavage site.
B, GLUT1 contains 197 potential α-chymotrypsin cleavage sites
(Phe, Tyr, Trp, Leu, Met, Ala, and Glu). The 52 detected cleavage sites are
indicated by  . Potential
α-chymotrypsin (α-CT) cleavage sites are present in all
TM domains. TMs are colored as in
Fig. 1.
. Potential
α-chymotrypsin (α-CT) cleavage sites are present in all
TM domains. TMs are colored as in
Fig. 1.