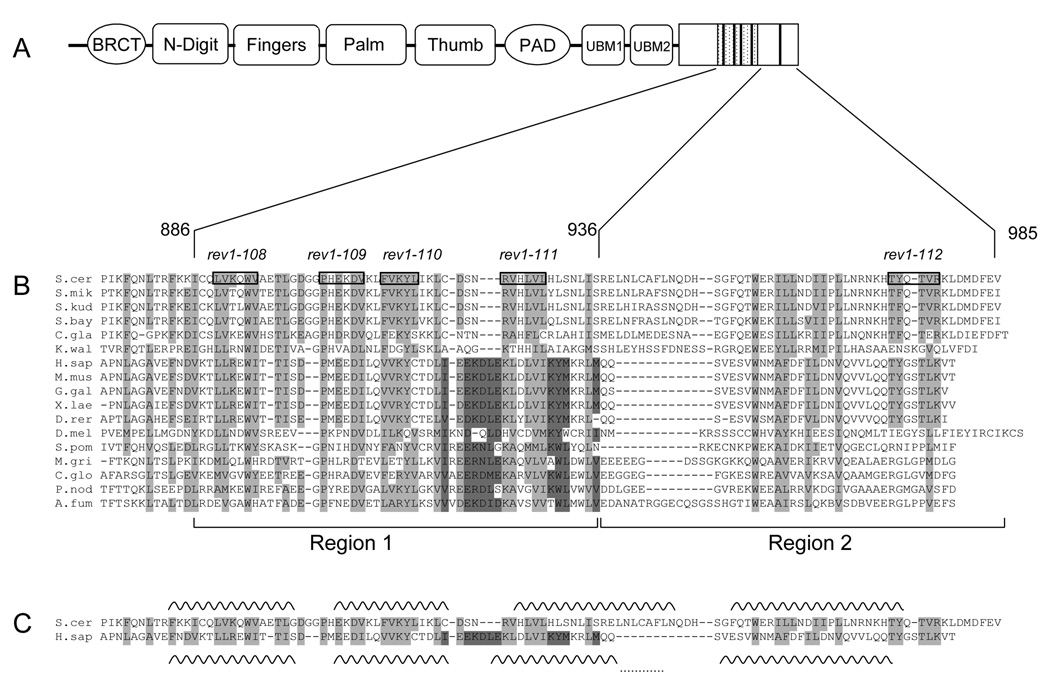
Fig. 1.
The C-terminal ~100 amino acids of Rev1 are conserved across a wide range of organisms. (A) Schematic of the domain structure of S. cerevisiae Rev1 with the minimal Rev7-interacting 50 amino acid Region 1 indicated by the hatched box and the conserved motifs indicated by dark bars. (B) Multiple sequence alignment of Rev1 sequences from selected species. Boxed residues and the text above indicate the alanine-patch mutations. (See Table 3 for more details). Amino acids highlighted in light grey show conservation across all species, while amino acids highlighted in dark grey indicate a conserved region not found in S. cerevisiae and closely related yeasts. Species abbreviation: S. cer, S. cerevisiae; S. mik, S. mikatae; S. kud, S. kudriavzevii; S. bay, S. bayanus; C. gla, C. glabrata; K. wal, K. waltii; H. sap, H. sapiens; M. mus, M. musculus; G. gal, Gallus gallus ; X. lae, X. laevis; D. rer, D. rerio; D. mel, D. melanogaster; S. pom, S. pombe; M. gri, M. grisea; C. glo, C. globosum; P. nod, P. nodorum; A. fum, A. fumigatus. Rev1 sequences from over 50 organisms were used to generate the alignment, but only 17 are shown due to space considerations. (C) Predicted secondary structure of the C-terminus of Rev1. Alignment of yeast and human Rev1 excerpted from (B) for clarity with the predicted helices for each sequence indicated by wavy lines. The dots connecting the last two helices of human Rev1 indicate that this helix is predicted to be continuous. (D) Helical wheel projections of the first predicted helix. Dark grey indicates hydrophobic residues, red indicates positively charged residues, blue indicates negatively charged residues and yellow indicates polar residues.