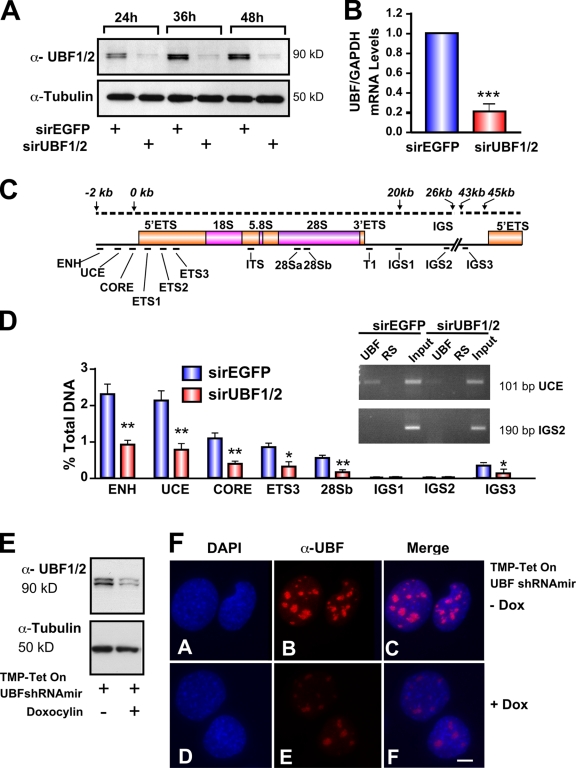
Figure 1.
Silencing of UBF1/2 by siRNA. (A) NIH3T3 cells were transfected with siRNA-EGFP or -UBF1/2, and protein samples were harvested as indicated and analyzed by Western blotting. (B) RNA samples were harvested, and UBF and GAPDH mRNA levels were examined by reverse transcription qRT-PCR. n = 3; ***, P < 0.001. (C) Schematic of a murine rRNA gene and the positions of qRT-PCR amplicons. (D) qChIP analysis of UBF binding to the rRNA gene. The percentage of DNA immunoprecipitated with anti-UBF or rabbit serum (RS) antibodies was calculated relative to the unprecipitated input control. The percentage of DNA of rabbit serum controls was subtracted from corresponding UBF samples. n = 6; *, P < 0.05; **, P < 0.01. A representative ethidium bromide gel shows the amount of UCE and IGS2 products amplified after 22 PCR cycles. (E) NIH3T3 cells stably transduced with tetracycline-inducible TMP-UBF shRNAmir were cultured in the presence or absence of doxocyclin and analyzed by Western blotting. (F) Samples in E were analyzed by immunofluorescence for UBF1/2. ENH, enhancer; ITS, internal transcribed spacer; T, terminator region. Mean ± SEM (error bars). Bar, 2 μm.