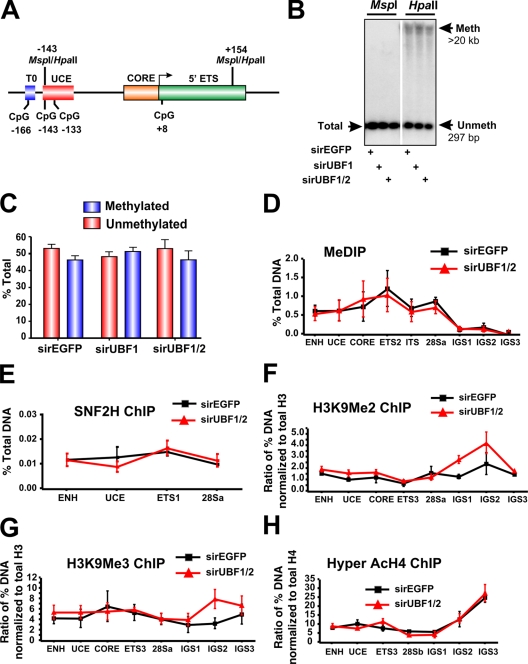
Figure 4.
DNA methylation and other heterochromatic markers are unaffected in response to UBF1/2 depletion. (A) Schematic of the murine rRNA gene promoter and 5′ ETS region with the position of restriction enzyme sites indicated. (B) Southern blot analysis of rDNA using methylation-sensitive restriction enzymes. Genomic DNA from an NIH3T3 cell transfected with siRNA-EGFP, -UBF1, or -UBF1/2 was digested with HpaII or MspI, subjected to electrophoresis, and hybridized to the probe depicted in Fig. 2 A. The white line indicates that intervening lanes have been spliced out. (C) The results (n = 2) in B were quantitated, and HpaII-sensitive (unmethylated) bands were quantitated as a proportion of the total rDNA (MspII band), and the difference was designated as methylated. (D) Analysis of DNA methylation across the rDNA by MeDIP in siRNA-EGFP or -UBF1/2 cells (n = 3). Samples were analyzed by qRT-PCR as described in Fig. 1 D. (E–H) Loss of UBF1/2 does not alter SNF2H binding or histone modifications associated with NoRC silencing of rDNA. qChIP analysis of the rRNA genes in siRNA-EGFP– or -UBF1/2–transfected NIH3T3 cells using antibodies against SNF2H (E), H3K9Me2 (F), H3K9Me3 (G), or hyperacetylated H4 (H). Samples were analyzed by qRT-PCR as described in Fig. 1 D. qChIPs (F–H) were normalized to total histone H3 or H4 loading (Fig. 6, A and B) and expressed as a ratio of the percentage of DNA (n = 3). ENH, enhancer; ITS, internal transcribed spacer; T, terminator region. Mean ± SEM (error bars).