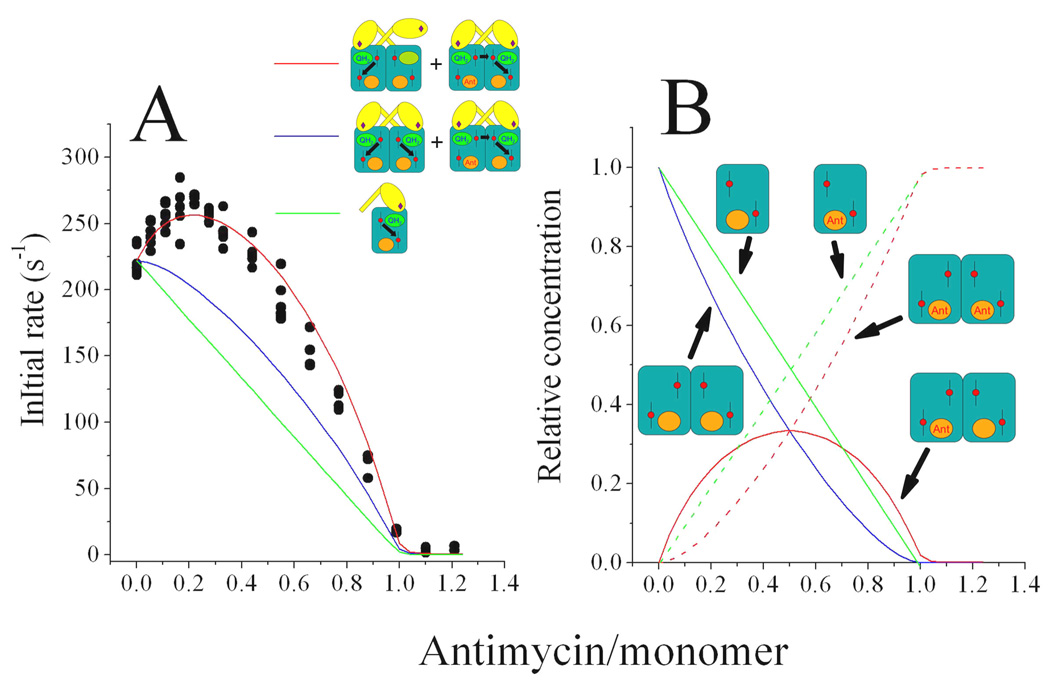
Fig. 12. Inhibition of the steady state ubiquinol-cytochrome c reductase activity of the yeast bc1 complex by antimycin.
Panel A shows data points (black circles) from four separate experiments in which the activity of 50 nM yeast bc1 complex was determined in the presence of various amounts of antimycin. The change in the activity as antimycin is added can be explained by assuming that only one monomer is active in the absence of inhibitor and that both monomers become active when only one center N is blocked (simulated by the red curve). Assuming that both monomers are active in the absence of antimycin and with one inhibitor per dimer (simulated by the blue curve) does not explain the experimental data, nor does the assumption that each monomer that binds antimycin becomes inactive (simulated by the green curve). Panel B shows the relative concentrations of free and inhibitor-complexed monomers and dimers as a function of antimycin concentration from which the curves in panel A were obtained: free monomers (green solid line), antimycin-bound monomers (green dashed line), unbound dimers (blue curve), dimers with one inhibitor (red solid curve) and dimers with two inhibitors (red dashed line). Data and simulations are from Ref. 15.