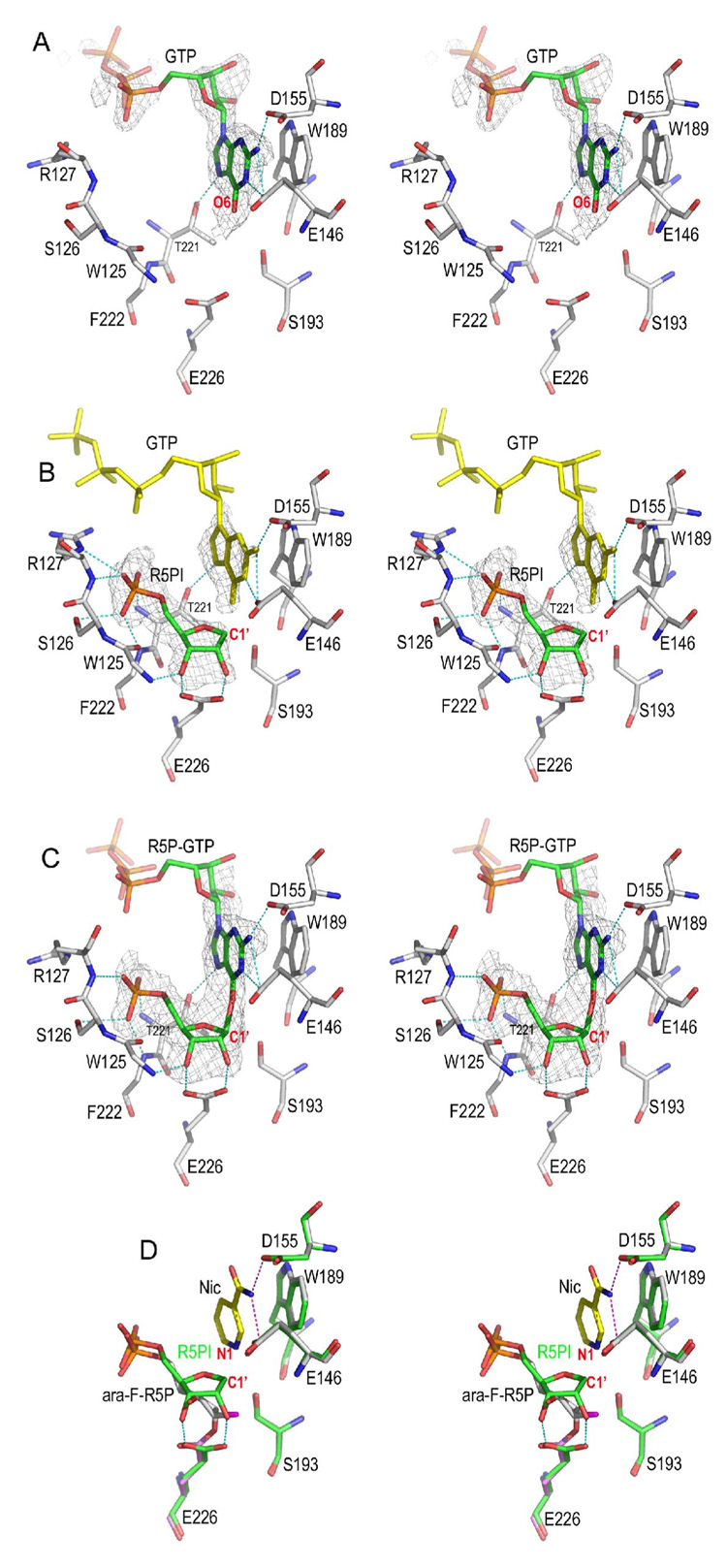
Figure 4. Non-covalent intermediate trapped by GTP.
A) The GTP complex. GTP alone can inhibit CD38 by occupying the nicotinamide binding site defined by residues Glu146, Asp155, and Trp189. The Fo-Fc omit (with GTP omitted from the calculation of the electron density map) electron density is shown as gray isomesh contoured at 2.5 σ covering GTP.
B) The non-covalent R5PI intermediate trapped by GTP. The non-covalent R5P intermediate (R5PI) is show as sticks with its carbon green. R5PI forms two hydrogen bonds with the catalytic residues Glu226 and Trp125 main chain nitrogen. The Fo-Fc omit electron density is shown as gray isomesh contoured at 2.8 σ. The GTP molecule in the active site (yellow sticks) has almost the same conformation as in the GTP complex shown in A). The phosphate group of R5PI forms extensive H-bonds to residues Ser126, Arg127, Phe222, and Thr221.
C) A R5P-GTP adduct in which R5PI is attacked by GTP from the β-face to form a covalent bond. The bond distance between C1′ and GTP O6 is 1.9 Å. The Fo-Fc omit electron density (with R5P-GTP omitted from the calculation of the electron density map) is shown as gray isomesh and contoured at 2.8 σ. D) Structural comparison of covalent and non-covalent intermediates. The covalent intermediate (green carbon sticks) with nicotinamide trapped in the active site is used for the superimposition with R5PI intermediate (gray carbon sticks). Essential active site residues Glu146, Asp155, Trp189, and Ser193 align quite well whereas the catalytic residue Glu226 rotates its carboxylate group by 30° in order to form a covalent intermediate with ara-F-R5P.