Abstract
Denitrifying microbial communities and denitrification in salt marsh sediments may be affected by many factors, including environmental conditions, nutrient availability, and levels of pollutants. The objective of this study was to examine how microbial community composition and denitrification enzyme activities (DEA) at a California salt marsh with high nutrient loading vary with such factors. Sediments were sampled from three elevations, each with different inundation and vegetation patterns, across 12 stations representing various salinity and nutrient conditions. Analyses included determination of cell abundance, total and denitrifier community compositions (by terminal restriction fragment length polymorphism), DEA, nutrients, and eluted metals. Total bacterial (16S rRNA) and denitrifier (nirS) community compositions and DEA were analyzed for their relationships to environmental variables and metal concentrations via multivariate direct gradient and regression analyses, respectively. Community composition and DEA were highly variable within the dynamic salt marsh system, but each was strongly affected by elevation (i.e., degree of inundation) and carbon content as well as by selected metals. Carbon content was highly related to elevation, and the relationships between DEA and carbon content were found to be elevation specific when evaluated across the entire marsh. There were also lateral gradients in the marsh, as evidenced by an even stronger association between community composition and elevation for a marsh subsystem. Lastly, though correlated with similar environmental factors and selected metals, denitrifier community composition and function appeared uncoupled in the marsh.
Salt marshes, located between uplands and open waters, often receive excessive amounts of nutrients (19) and accumulate pollutants, such as toxic metals (47, 67). Salt marshes are recognized for their potential to intercept and prevent nutrient loading to receiving waters (28, 55), in part due to their diverse and active microbial communities (7, 31), which catalyze processes such as denitrification, i.e., a major pathway for alleviation of eutrophication in coastal waters (28, 75). However, salt marshes are complex and dynamic systems, with environmental conditions varying extensively in space and time (2), and thus, denitrification studies with salt marsh systems have often focused on temporal and spatial patterns of the process (41, 71) and effects of environmental factors, such as temperature, oxygen, salinity, and availability of nutrients (34, 54). Yet, denitrifying bacterial communities, which are also shaped by environmental factors and pollutants, are the biological transducers through which these factors influence the denitrification process (76). As such, soil studies have shown that denitrifier community composition can regulate denitrification (14, 27). For salt marsh sediments, however, the relationships between denitrifier community composition and denitrifier function are less understood. Also unknown are the way that the relationships vary spatially within the marsh and the relative importance of environmental factors that also vary spatially, such as inundation, nutrient content, and levels of potentially influential pollutants.
Though denitrifier communities in soil (5, 49, 51, 64, 77) and in marine sediments (8, 10, 45, 53, 68), where they may be influenced by season, nutrient concentration, soil types, redox gradients, and fertilizer types, have been widely studied, those in salt marsh systems have received little attention. Salt marshes differ fundamentally from soils and marine sediments in that marsh sediments are tidally influenced with associated redox potential, salinity, and inundation fluctuations (50). Studies of total bacterial communities in salt marsh sediments indicate that while they are highly spatially variable, they are significantly influenced by geographic location, elevation, and selected pollutants, such as metals (12, 18). Yet, rhizosphere diazotroph communities were shown to be relatively stable and persistent (46) in the face of short-term carbon and nutrient variability (61, 62). How denitrifier communities and their functions may vary with environmental and pollutant factors in salt marshes remains largely unknown. Therefore, the objective of this study was to examine, for a salt marsh system with high nitrate loading (57), the spatial gradients of microbial community composition and denitrification enzyme activities (DEAs), the relationships of community composition and function to environmental factors as well as to each other, and the potential effects of metals considered here to be sedimentary pollutants that could affect denitrifiers and denitrification (12, 18).
MATERIALS AND METHODS
Site description.
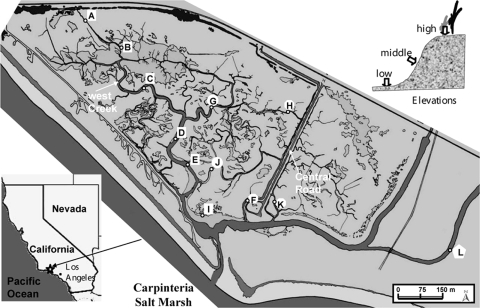
The study site was the Carpinteria Salt Marsh (CSM) (Fig. 1), a 93-ha estuarine wetland located 12 km east of Santa Barbara, CA (34°24′N, 119°31′W), with West Creek flowing through the western marsh area. The region has a semiarid climate typical of southern California, with annual average temperatures ranging from 10.2 to 21.7°C and an annual average rainfall of 44.7 cm, 92% of which falls from November through April. The CSM has historically received elevated levels of nitrate from upland agricultural activities (57).
FIG. 1.
Study location including 12 sampling stations. The stations are divided into four groups: stations along the main channel of West Creek (A, B, C, D, E, and I), stations on branches of West Creek (G and J), stations along the central road (H, F, and K), and a station further east of the marsh (L).
Sample collection.
Twelve stations (Fig. 1) were sampled on 26 August 2003. At each station, sediment samples were taken from three elevations (Fig. 1): high (marsh edge, usually vegetated), middle (creek bank), and low (channel bottom). Sediment samples were collected by pushing a piston core, constructed from a 60-ml disposable polypropylene syringe (B-D, Franklin Lakes, NJ), into the sediment while maintaining minimal pressure to avoid overcompaction. Ten to 12 individual cores were collected within a 1-m-diameter area at each sampling site, and the top 2.5 cm was pooled into sterile Qorpak jars (Fisher Scientific, Tustin, CA) and homogenized by stirring. A prior experiment determined that DEA was preferentially expressed in the surface 2.5-cm layer (data not shown). Bulk water samples were collected immediately above the sediment surface before sediment sample collection. Water and sediment samples were immediately transported on wet ice to the laboratory, where they were processed at 4°C within 24 h.
Sediment characterization.
Moisture content and total organic carbon (TOC) were measured for all 36 samples. Moisture content was determined for triplicate sediment samples (1.5 g each) according to standard methods (23). Dried (105°C; O/N) sediments were combusted in a muffle oven at 450°C to determine TOC (52). The remaining sediments were preserved at −20°C until microbial community and sediment eluate analyses (as described below).
Dissolved nutrient analysis.
Bulk water samples collected from surfaces immediately above the sediment at the low elevations were analyzed for concentrations of nitrate (38), phosphate (as soluble reactive phosphorus [39] and ammonium [37]), using a model 2300-000 ion analyzer (QuikChem II; Lachat, Milwaukee, WI). Before nutrient analysis, the water samples were filtered through a filter sandwich (0.7-μm Whatman GF/F filter overlaying a 0.2-μm Millipore filter) to eliminate the suspended particles and microbes in the samples. Filtered water samples were stored at −20°C until analysis (57).
Sediment eluate analyses.
Bioavailable metals in sediment eluates were quantified. To recover eluate, 15 g of thawed and homogenized sediment was weighed into 50-ml polypropylene centrifugation tubes and 15 ml chilled (4°C) sterile artificial seawater (ASW) was added. After an initial brief vigorous shaking to ensure thorough mixing, samples were horizontally shaken (2 h, 4°C) and then centrifuged (2500 × g, 30 min, 4°C). Eluted water samples were collected and analyzed for the concentrations of metals and dissolved organic carbon (DOC). Eluates were similarly collected and analyzed for additional fresh sediment samples to determine the effect of the sediment freeze-thaw process on DOC and metal concentrations. For all samples, analysis controls included either ASW or ultrapure water (carbon and ion free) in either centrifuge tubes or combusted (450°C; O/N) glass vials.
An aliquot of each eluate (250 μl) was diluted 50-fold in 1% nitric acid (trace metal grade; Fisher Scientific, Pittsburgh, PA) in a new polypropylene centrifuge tube. A blank tube was measured as a control, confirming negligible contamination. Metal quantification was performed by direct aspiration using an inductively coupled plasma mass spectrometer (model 7500i; Agilent, Palo Alto, CA) and compared to external standards, covering a range from 0.01 μg l−1 to 200 μg l−1 for minor elements and up to 100 mg l−1 for major elements. Analysis was performed as prescribed by EPA method 6020 (74). The instrument was operated with very low-charge molecular ions, a robust plasma characterized by 0.4% CeO/Ce. For DOC, eluates were diluted 100-fold and analyzed via high-temperature combustion, using a Shimadzu TOC-V instrument (Shimadzu Scientific Instruments, Columbia, MD), as described previously (13). Three to five replicate 100-μl samples were injected into the combustion tube for analysis. The injections continued until at least three injections met the specified levels (a standard deviation of 0.1 area counts and a coefficient of variation of ≤2%); otherwise, the best three of five injections were considered. Both DOC and metal concentrations were corrected for analysis controls and normalized to sediment dry weight.
DEA.
DEA was measured using a standard method (72) for triplicates of each sediment sample within 36 h of sample collection. Briefly, ca. 3 g of sediment was placed in a 15-ml sterile, airtight, Hungate-type anaerobic culture tube (Bellco Glass, Inc., Vineland, NJ) containing 1 mM glucose, 1 mM KNO3, and 1 g l−1 chloramphenicol. The headspace was flushed with 7 volumes of filter-sterilized (0.2 μm) helium. Acetylene (10% in He) was added with a syringe through the septum, and the sediment slurry was incubated on a rotary shaker (30°C, 2.5 h). During the incubation, the headspace was sampled (3 ml) and replaced three times with 10% acetylene at 0.5, 1.5, and 2.5 h. The headspace concentration of N2O was determined by gas chromatography with an electron capture detector (GC-14A; Shimadzu Corporation, Columbia, MD), using a chromatography laboratory automated software system (CLASS-VP; Shimadzu Corporation, Columbia, MD) from a calibration curve generated with N2O standards (Scott Specialty Gases, Inc., Plumsteadville, PA). The above-mentioned sampling and incubation times were determined experimentally to achieve the best linearity of DEA for the sediment samples (72).
Abundance of bacterial cells.
Enumeration of total bacterial cells was performed in triplicate, using a method similar to that of Holden et al. (25). Briefly, 150 μl of the sediment suspension was stained with a nucleic acid-specific stain (Sybr gold; Molecular Probes, Eugene, OR) and then suction filtered onto a 13-mm-diameter, 0.2-μm-pore-size black Isopore membrane (Millipore, Bedford, MA). Total cells were directly counted using a Nikon E800 epifluorescence microscope with a green fluorescent protein filter set (Chroma, Brattleboro, VT); counts were normalized to sediment dry weight.
Microbial community analysis.
The total bacterial and denitrifier communities were profiled via terminal restriction fragment length polymorphisms (TRFLPs) as described previously (40). Briefly, 2 g of sediment was extracted using an UltraClean soil DNA kit (MoBio, Solana Beach, CA), following the manufacturer's protocol. Extracted DNA was purified (by size exclusion with Sepharose spin columns) and checked for quality using A260/230 spectrophotometry and for quantity using Picogreen (Molecular Probes, Eugene, OR) fluorometry. For profiling of the total bacterial community, genes encoding 16S rRNA were PCR amplified from purified community DNA by using universal eubacterial primers 8F hex (fluorescently labeled forward primer; 5′AGAGTTTGATCCTGGCTCAG) (44) and 1389R (5′ACGGGCGGTGTGTACAAG) (56). For the denitrifying community, genes encoding the nitrite reductase NirS were PCR amplified using the primers nirS1F hex (fluorescently labeled forward primer; 5′CCTAYTGGCCGCCRCART) (9) and nirS6R (5′CGTTGAACTTRCCGGT) (9) as described previously (10). PCR products were purified and digested using either HhaI or MspI in two separate digestions (21). The restriction enzymes were inactivated (65°C, 20 min). The lengths of fluorescently labeled terminal restriction fragments (TRFs) were determined with an Applied Biosystems Instruments (ABI; Foster City, CA) model 373A automated sequencer. Data from HhaI and MspI digestions were separately aligned and normalized using a method similar to that of Dunbar et al. (20) and then concatenated to form a collective data set. All TRFLP data were analyzed in binary form (i.e., presence and absence of TRFs) unless otherwise indicated. Data analyses using relative abundances of TRFs (i.e., normalized to the total fluorescence peak height) were conducted for comparison. Numbers of TRFs were tabulated for a comparative measure of community richness.
Data analyses.
All data analyses were conducted using either S-plus version 6.1 (Insightful Corp., Seattle, WA), Primer version 6 (PRIMER-E Ltd, Plymouth, United Kingdom), or CANOCO version 4.53 (Microcomputer Power, Inc., Ithaca, NY). Continuous variables (organic content and metal concentrations) were standardized to zero mean and unit variance before either regression or multivariate correlation analyses in order to account for differences in the values of these variables. The correlation structure among metal concentrations was determined by calculating pairwise Pearson's correlation coefficients with S-plus. Effects of elevation on DOC, TOC, and metal concentrations were evaluated via analysis of variance. The relationship between any two continuous variables was investigated by simple regression analysis. To investigate the effects of metals on a single biological variable (e.g., DEA and total cell counts, as opposed to multivariate community profiles) in the presence of other environmental variables (such as DOC, TOC, and elevation), the residual variability in either DEA or total cell counts (after adjustment for carbon and/or elevation effects) was correlated with concentrations of specific metals (36) with S-plus. Bray-Curtis and Euclidean distances were calculated for microbial community TRFLP profiles and for abiotic characteristics (i.e., environmental variables and metal concentrations), respectively, to summarize their relative spatial distributions in the marsh. Analyses of correlation between these distance measures were then conducted through the RELATE program in Primer version 6 (16) to compare, for example, the spatial distributions of microbial communities in the 16S TRFLP and the nirS TRFLP profiles for all samples or at each elevation. The Spearman coefficient (32) was used as a measure of correlation, and P values were calculated via permutation.
Three multivariate techniques based on correspondence analysis were employed to analyze the TRFLP profiles (58, 69, 70). Detrended correspondence analysis (DCA) was used to characterize the overall variation in the TRFLP profiles. Canonical correspondence analysis (CCA) was used to directly assess the relationships between microbial community profiles and environmental variables such as elevation, TOC, and DOC, with dummy variables included to account for sample elevation. CCA was also used to examine relationships between TRFLP profiles and other biotic measures, such as DEA and total cell counts. A third technique, partial CCA (pCCA), was used to investigate the effects of metals on microbial communities after adjustment for natural variability due to elevation and carbon content. Briefly, variables representing elevation and carbon content were designated covariables, and the residual variabilities in the TRFLP profiles were correlated with metal concentrations, as previously described (12).
RESULTS
Nutrient concentrations.
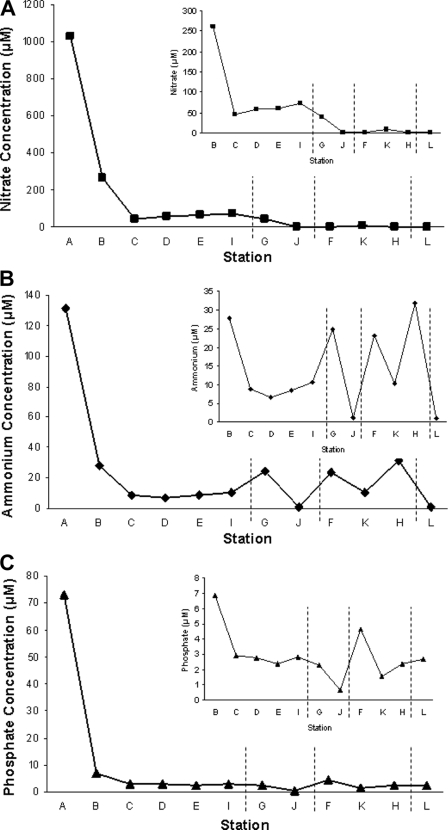
The most upstream station, station A, had the highest water column concentrations of dissolved nitrate, ammonium, and phosphate among all 12 stations (Fig. 2). There appeared to be trends of decreasing concentration downstream along the main channel of West Creek (Fig. 2, stations A, B, C, D, E, and I). Of the two stations on the West Creek branches, station G was more upland and had higher nutrient concentrations than station J, downstream. Stations along the central road (F, K, and H) and further east of the marsh (L) showed no obvious pattern regarding nutrient concentrations.
FIG. 2.
Dissolved nutrient concentrations in bulk water samples overlying low-elevation sampling sites at each station: nitrate (A), ammonium (B), and phosphate (C). Concentrations were based on single-sample measurements. Inset graphs exclude station A to emphasize differences across the remaining stations. Vertical dashed lines separate the groups of stations described in the legend to Fig. 1.
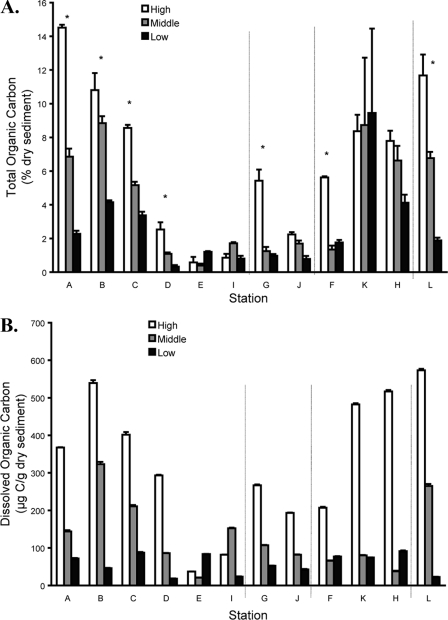
Sediment TOC (Fig. 3A) and DOC (Fig. 3B) concentrations were greater at high elevations than at middle and low elevations for most stations, and there was a trend of decreasing concentrations for high elevations from upstream stations to downstream stations along West Creek. Across all stations and all elevations, both DOC (R2 = 0.51; P = 0.0000076) and TOC (R2 = 0.19; P = 0.030) were significantly related to elevation. TOC and DOC were auto-correlated (R2 = 0.54; P < 0.0000001), and approximately 0.06% to 1.16% of TOC was soluble, with larger percentages in samples from high elevations than in those from either middle or low elevations (R2 = 0.17; P = 0.044). Freeze-thaw preparations of preserved sediments increased DOC in sediment eluates, compared to the level for fresh sediments, to the same degree for different samples (data not shown). Because DOC and TOC were highly correlated, and because DOC was measured using the sediment eluates for which metal concentrations were determined, DOC rather than TOC was used for comparison to other data in this study (e.g., DEA and community profiles, etc.).
FIG. 3.
TOC (A) and DOC (B) for three elevations (high, middle, and low) at each station. Vertical dashed lines separate the groups of stations described in the legend to Fig. 1. Asterisks in panel A denote stations where TOC differed significantly across elevations. DOC differed significantly across elevations in all 12 stations. The bars represent the mean values, and the error bars represent the standard deviations (n = 3).
Metals in sediment eluates.
Twenty-six metals in the sediment eluates were analyzed, and the freeze-thaw process had no significant effect on metal recovery in the eluates (data not shown). Sodium concentrations (Table 1) in sediments, after adjustment for that in ASW, were used as a proxy for in situ salinity. Concentrations of metals varied widely (Table 1), without respect to either elevation or station, and were comparable to those reported for a previous year for this marsh (12). Thus, metal concentrations at individual stations and elevations are not distinguished in Table 1. Also, of the total 26 metals detected, 5 (Ag, Be, Cd, Cs, and Tl) had very low concentrations that were indistinguishable from those in ASW and thus were neither reported in Table 1 nor included in statistical analyses. The concentrations of many of the 26 analyzed metals were highly correlated, and 14 metals (Al, Ba, Ca, Cr, Co, Cu, Li, Mn, Ni, Pb, Se, U, V, and Zn) were selected as “representative” to reduce the complexity of the variable matrix for analysis of relationships to microbial community measurements. The following high correlations (Pearson's correlation coefficient ≥ 0.85) existed between the concentrations of the representative metals and the remaining metals: Al-Fe, 0.94; Al-Rb, 0.89; Al-As, 0.88; Ca-Mg, 0.95; Ca-Sr, 0.94; Ca-K, 0.92; Ca-Rb, 0.86; Ca-As, 0.85; Ca-Fe, 0.88; Cr-Mg, 0.90; Cr-K, 0.89; Cr-Rb, 0.89; Cr-As, 0.88; Li-Mg, 0.94; Li-K, 0.88; Ni-As, 0.96; Ni-Rb, 0.86; Ni-Sr, 0.85; Se-Mg, 0.91; Se-K, 0.91; and Se-Rb, 0.87. Also, many of the concentrations of representative metals were well correlated: Ca-Se, 0.86; Ca-Ni, 0.87; Ca-Cr, 0.85; Ca-Li, 0.85; Cr-Li, 0.87; Cr-Ni, 0.87; Cr-Se, 0.86; and Cr-Ca, 0.85. Cu and Ni were also correlated, albeit comparatively weakly (0.80). Among the 14 representative metals, Zn was significantly positively related to both DOC (R2 = 0.13; P = 0.030) and TOC (R2 = 0.18; P = 0.010), and V was significantly positively related to DOC (R2 = 0.11; P = 0.047).
TABLE 1.
Metal concentrations in sediment eluatesa
| Metal | Concn range | Mean | SD |
|---|---|---|---|
| Na | 4.1-21.4 | 10 | 4.6 |
| Ca | 0.1-0.7 | 0.4 | 0.2 |
| K | 0.2-0.9 | 0.4 | 0.2 |
| Mg | 0.46-2.4 | 1.2 | 0.5 |
| Al | 48.1-1,824.5 | 534.0 | 529.5 |
| As | 18.6-172 | 70.7 | 44.7 |
| Ba | 7.5-165.8 | 60.7 | 41.3 |
| Co | 2-22.8 | 7.7 | 5.2 |
| Cr | 7.8-60.5 | 28.6 | 12.0 |
| Cu | 0-276.9b | 48.3 | 68.3 |
| Fe | 681.5-4,199.9 | 1,827.6 | 910.5 |
| Li | 115.3-774.2 | 361.2 | 149.3 |
| Mn | 0-8,831.2b | 725.4 | 1,659.9 |
| Ni | 30.5-279.3 | 97.6 | 65.7 |
| Pb | 0-10b | 1.5 | 2.5 |
| Rb | 56.1-339.8 | 153.9 | 67.5 |
| Se | 24.6-121 | 67.7 | 26.4 |
| Sr | 2,722.1-11,440.7 | 6,066.6 | 2,377.7 |
| U | 2.6-122.5 | 26.5 | 25.7 |
| V | 6.5-67.7 | 23.5 | 15.1 |
| Zn | 20.1-384.6 | 83.0 | 77.6 |
Values for Na, Ca, K, and Mg are given in mg g−1 dry sediment, and those for the rest of the elements are given in ng g−1 dry sediment. Means are averages for 36 samples.
Minimum concentrations were less than or similar to background concentrations in ASW.
Microbial community profiles.
Microbial community profiles were obtained for both the total bacterial and the denitrifier communities, using 16S and nirS TRFLPs, respectively, and the numbers of TRFs were counted as richness estimates. Overall, the total bacterial and denitrifier profiles had similar magnitudes of TRFs, and averaged across stations, the number of 16S-TRFs decreased while the number of nirS-TRFs increased from the marsh edge to the creek bank and the channel bottom (Table 2). Spatial variability was higher for denitrifier than total bacterial community profiles (by DCA; see below), yet the spatial distributions of both communities were marginally similar (Table 3). By DCA, the variation in community profiles, a.k.a. inertia, was 1.6 times higher in nirS TRFLP profiles (inertia, 4.32) than in 16S TRFLP profiles (inertia, 2.72), indicating that the subcommunity (denitrifiers) was more spatially variable than the overall community (total bacteria). Yet, the positive correlation coefficients found upon comparison of 16S and nirS TRFLP profiles, both throughout the marsh (ρs = 0.21) (Table 3) and along the 12 stations at the middle elevation (ρs = 0.35) (Table 3), suggest that the average trends in spatial distribution throughout the marsh were similar for the two communities.
TABLE 2.
Numbers of TRFs in the 16S TRFLP and nirS TRFLP profiles
| Profile and area | No. of TRFsa
|
||
|---|---|---|---|
| Avg | Minimum | Maximum | |
| 16S TRFLP | |||
| Marsh edge | 51 | 40 | 66 |
| Creek bank | 45 | 31 | 59 |
| Creek bottom | 43 | 25 | 54 |
| nirS TRFLP | |||
| Marsh edge | 37 | 22 | 49 |
| Creek bank | 47 | 32 | 60 |
| Creek bottom | 43 | 30 | 67 |
Data for all 12 stations are averaged (“avg” column), and the ranges of TRF abundances are indicated in the “minimum” and “maximum” columns.
TABLE 3.
Correlations between 16S and nirS TRFLP community profiles for all samples at each elevation
ρs is the Spearman rank coefficient of correlation between the 16S and nirS TRFLP profiles. This coefficient is calculated based on the Bray-Curtis distances of the compared community profiles (16). A high ρs (≤1 here) indicates a high degree of similarity between the spatial distributions of the total bacterial and the denitrifier communities throughout the marsh (all elevations) or along the 12 stations for each elevation (high, middle, and low).
A P value of <0.05 indicates a significant ρs.
The 16S TRFLP and nirS TRFLP profiles for the high elevation were significantly different from those for the middle and low elevations (P = 0.002 and P = 0.028 for 16S and nirS TRFLP profiles, respectively), but the nirS TRFLP profiles from the low and middle elevations were similar (P = 0.15). The 16S TRFLP and nirS TRFLP profiles were also significantly correlated with DOC (P = 0.002 and P = 0.012, respectively) and TOC (P = 0.006 and P = 0.056, respectively). By CCA, significant amounts of the total variations in the 16S TRFLP (13%; P = 0.002) and in the nirS TRFLP (10%; P = 0.034) profiles were jointly explained by three variables: high elevation, low elevation, and DOC. Also, when a subsystem (comprising stations A, B, C, and D, along the main channel of West Creek) within the whole marsh was analyzed, elevation was found to be more important, explaining 13% and 21% of the overall variations for 16S and nirS TRFLP, respectively, compared to 10% and 6% of the overall variations for the whole marsh (all 12 stations) (Fig. 1). However, the amounts of variation in community composition explainable by DOC were found to be similar when both the subsystem and the whole marsh were evaluated. No additional variation in either the 16S or the nirS TRFLP community profiles was explained by sodium concentration (a proxy for salinity).
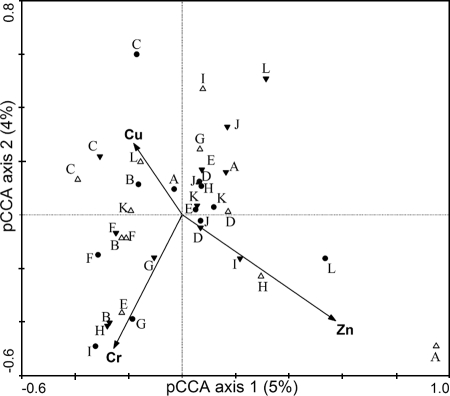
Because significant natural variability (i.e., related to elevation and carbon content) was present in the microbial community profiles, three variables (high elevation, low elevation, and DOC) were included as covariables for correlating metal concentrations with the community profiles via pCCA. The relationships of metals to community profiles were determined in three ways: by forward selection, by manual selection, and by correlation with metals that were selected by the previous two methods. More emphasis is placed on relationships determined by the first two methods because those metals were directly correlated with community composition while those in the last category were inferred. Via automated forward selection in pCCA, three metals explained the residual variation in the 16S TRFLP profiles: chromium (Cr), zinc (Zn), and copper (Cu) (Fig. 4). Each of the 14 metal pollutant variables was also tested manually for a correlation (i.e., ability to explain the residual variation) with 16S TRFLP profiles, using the Monte Carlo permutation test, and Cu (P = 0.075), Cr (P = 0.081), Ni (P = 0.066), and Li (P = 0.083) were marginally correlated in the presence/absence of 16S TRFLP data but significantly (P < 0.05) correlated in a relative abundance of 16S TRFLP data. Se, which was highly correlated with Cr (0.86), was marginally correlated in a relative abundance of 16S TRFLP data (P = 0.07). Mg, Rb, As, and K, which were not included in pCCA as representative metals, may also be related to 16S TRFLP profiles due to their high correlation with Cr (>0.85; see above).
FIG. 4.
Ordination diagram from pCCA of 16S TRFLP data, where elevations and DOC were used as covariables. Metals are represented by arrows, the lengths of which indicate the strength of correlation between the metal and the community data. Samples are labeled using their station names and denoted by triangles and circles. Open triangles pointing upward indicate high-elevation samples, filled triangles pointing downward indicate low-elevation samples, and filled circles indicate middle-elevation samples.
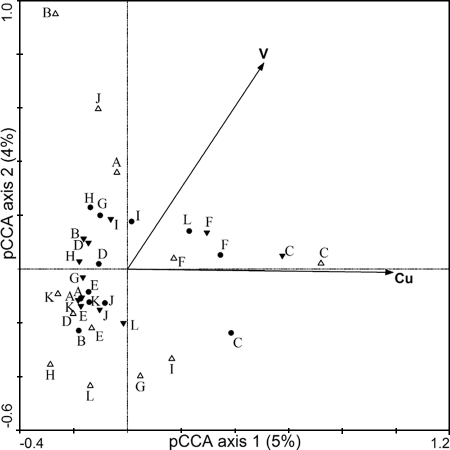
Similar to the results for pCCA of 16S TRFLP profiles, after adjustment for natural variability, two metals were found to best explain the residual variation in the nirS TRFLP profiles: vanadium (V) and Cu (Fig. 5). Each metal variable was also tested manually for its correlation with nirS TRFLP profiles by using the Monte Carlo permutation test, and nickel (Ni; P = 0.032), selenium (Se; P = 0.020), Cr (P = 0.020), and Cu (P = 0.018) were significantly correlated with nirS TRFLP profiles. As above, because As, Mg, Rb, and K, which were not included in pCCA as representative metals, are highly correlated with Cr and/or Se and/or Ni (>0.85; see above), they may also be related to nirS TRFLP profiles. In summary, Cu, Cr, and Ni appeared to influence both the 16S and the nirS profiles, while Zn was additionally related to 16S profiles and V was additionally related to the nirS profiles. Other metals (i.e., As, Mg, Rb, Li, Se, and K) were correlated with many of these metals and thus may have either a direct or an indirect relationship to the community profiles.
FIG. 5.
Ordination diagram from pCCA of nirS TRFLP data, where elevations and DOC were the covariables. Metals are represented by arrows, the lengths of which indicate the strength of correlation between the metal and the community data. Samples are labeled using their station names and denoted by triangles and circles. Open triangles pointing upward indicate high-elevation samples, filled triangles pointing downward indicate low-elevation samples, and filled circles indicate middle-elevation samples.
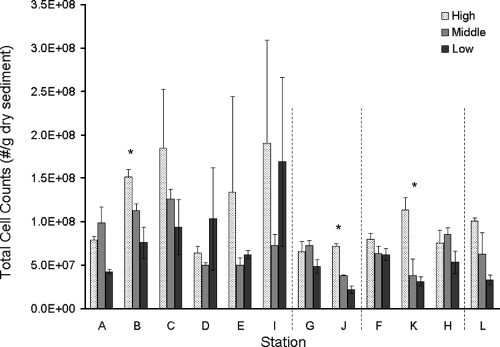
Total cell counts.
Total cell counts were relatively similar across stations throughout the marsh and were generally higher (R2 = 0.20; P = 0.025) at high elevations than at middle and low elevations (Fig. 6). Across all stations, total cell counts were positively correlated with DOC (R2 = 0.12; P = 0.043) but not with TOC and appeared negatively, but was not significantly, related to sodium concentration. When correlations between total cell counts and DOC for each elevation were analyzed separately, a significant correlation was found only for the middle elevation (R2 = 0.36; P = 0.040). Total cell counts were negatively correlated with concentrations of several metals (V [P = 0.013], Cr [P = 0.046], and Cu [P = 0.025]), but only the relationship to Cu was significant after adjustment for DOC effects. Correlations of total cell counts with microbial community profiles were also examined using CCA: total cell counts were significantly correlated with 16S TRFLP patterns (P = 0.03) but not with nirS TRFLP patterns (P = 0.12).
FIG. 6.
Total cell counts. Vertical dashed lines separate the groups of stations described in the legend to Fig. 1. Asterisks denote stations where total cell counts differed significantly across elevations. The bars represent the mean values, and the error bars represent the standard deviations (n = 3).
DEA.
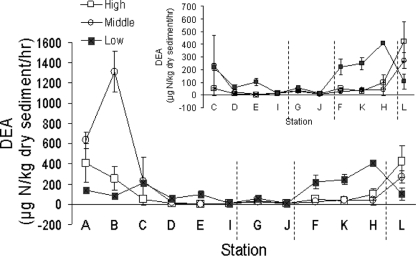
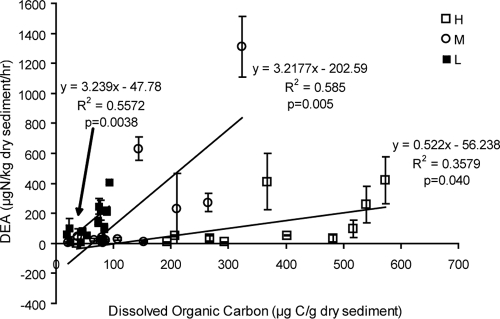
DEA is affected by the population sizes of denitrifiers, the levels of expression of the denitrification genes, and the efficiency of denitrification enzymes, which are likely influenced by denitrifier community composition. Overall, DEA was highly spatially variable in the marsh and did not consistently vary with either station or elevation (Fig. 7) but increased with DOC (Fig. 8) and was correlated with several metals. Stations upstream of West Creek, along the central road, and further east of the marsh appeared to be DEA “hot spots,” and the sediment sample from the creek bank of station B had an especially high DEA (Fig. 7), probably due to the high organic C (Fig. 3) and nitrate (Fig. 2A) concentrations. Across all stations, DEA was significantly related to DOC, but DEA increased more rapidly with increasing DOC at the low (and middle) elevations than at the high elevations (Fig. 8). Two high DEA values for the middle elevation (Fig. 8) appeared as outliers, and without those two points, the relationships between DEA and DOC were found to be similar when the middle and high elevations were compared. Because DOC and TOC were highly correlated (see above), DEA was also significantly related to TOC. DEA appeared to be higher at stations A and B, where dissolved nutrients were also relatively higher (Fig. 2 and 7), but overall DEA did not vary significantly with nutrient concentrations.
FIG. 7.
DEA measurements for three elevations (high, middle, and low) at each station. The inset graph excludes stations A and B to facilitate display. Vertical dashed lines separate the groups of stations described in the legend to Fig. 1. The data points represent the mean values, and the error bars represent the standard deviations (n = 3).
FIG. 8.
Relationship between DEA and DOC. Overall regression analysis across all three elevations yielded R2 values of 0.092 and P values of 0.072. The data points represent the mean values, and the error bars represent the standard deviations (n = 3).
Across all stations and elevations in the entire marsh, as well as within the subsystem defined along stations A, B, C, and D, DEA was not correlated (by CCA) with either 16S TRFLP or nirS TRFLP profiles. However, when analyzed for each elevation separately, DEA appeared to be somewhat related to nirS TRFLP at low elevations (P = 0.14) but not at middle (P = 0.72) or high (P = 0.90) elevations.
When the variables of elevation and carbon were included in the statistical analyses, DEA was not significantly correlated with either the sodium concentrations or the metal levels. Therefore, to specifically evaluate the effects of metals on DEA, the variability residuals remaining after adjustment for the elevation-dependent DOC effects were correlated with metal concentrations. Four metals then appeared to be significantly negatively correlated with DEA: Pb (P = 0.0060), Al (P = 0.0068), Ni (P = 0.038), and Ba (P = 0.047). As, Fe, Ca, Rb, and Sr may also be negatively related to DEA due to their high positive correlations with Ni and/or Al (correlation coefficients of >0.85; see above).
DISCUSSION
Though highly variable in the dynamic salt marsh system, both denitrifier community composition and DEA were affected by similar factors, such as elevation, carbon content, and concentrations of specific metals. The influence of DOC on DEA varied significantly with elevation across the whole marsh. Elevation affected community composition across the entire marsh, and the effect was further magnified within the West Creek subsystem, indicating that, in addition to variations by elevation, there were also variations along lateral gradients in the marsh. Nevertheless, denitrifier community composition and function appeared to be uncoupled.
The nirS communities were highly variable, more so than the 16S communities, and appeared to be subjected to multiple gradients or factors in the salt marsh. Similarly, a previous study also showed that functional genes with faster evolutionary rates, such as nirS, had higher levels of diversity than the more conserved genes encoding 16S rRNA (8). In this study, elevation appeared to account for the extensive variations in microbial community composition. This was similar to what was found in one of our previous studies (12) but different from what was found in another study regarding TRFLP profiles across several marshes (18). The contrast occurs because prevailing explanatory variables will change depending on the spatial and temporal context of a study. For example, in Córdova-Kreylos et al. (18), six marshes at different latitudes were analyzed in multiple years, and marsh, latitude, and year were the strongest factors influencing TRFLP profiles. On the other hand, Cao et al. (12) found that elevation was a significant factor influencing TRFLP profiles when only two marshes from one single year were studied. In a retrospective analysis of the latter study, performed here for comparison, the effect of elevation became even stronger when the differences between the two marshes (i.e., the “marsh” gradient) were statistically removed, i.e., elevation then explained 27% of the variation in 16S TRFLP profiles for one marsh (Fig. 1, stations A, B, C, and D), compared to 15% of the variation explained when both marshes were considered (12). Even when only one marsh is of concern (in this study), the differences between the subsystems (West Creek, the central road, and the east marsh) inside the single marsh are also potentially influential.
DEA was also highly variable within the marsh and was significantly correlated with DOC, but the correlations differed among elevations. The DEAs in this study were of magnitudes similar to those in prior reports, i.e., for a riparian zone affected by swine wastewater (3 to 1,660 μg N2O-N kg−1 dry soil h−1) (29), a rocky estuarine biofilm (140 to 840 μg N2-N kg−1 wet weight h−1) (48), and for salt marsh sediments amended with nitrate (1,500 μg N2O-N kg−1 dry soil h−1) (1). These, in turn, were all slightly higher than the levels for intertidal sediments (14 to 126 μg N2-N kg−1 wet weight h−1) (48). DEA is a measure of denitrifier abundance, denitrification pathway gene expression levels, and denitrification enzyme efficiencies. While the significant correlation between DEA and DOC in this study was consistent with the general knowledge (72), as most denitrifiers are heterotrophs (33) and often C limited (11, 15, 59), the correlations varied within the salt marsh, where multiple gradients or patchiness exist. For example, when samples from the three elevations, with their differing characteristics, were analyzed together, DEA was only marginally related to DOC (P = 0.072) and only 9.2% of DEA variability could be explained (Fig. 8). In contrast, when each elevation was analyzed separately, not only could a larger percentage of variability (on average, 50%) in DEA be explained by DOC, but also DEA in high elevations was much less responsive to increased C. This suggests an interaction between the elevation gradient and the DOC gradient. The interaction may result from factors affecting denitrification, such as competition of marsh plants for nitrogen (61), quality (61) and abundance (15) of DOC, and rhizosphere influences that can dampen the effects of environmental variations on microbes (6). These factors are part of the elevation gradient but perhaps vary inconsistently across the 12 stations. Also, because other factors, such as nitrate concentration, salinity, temperature, pH, oxygen supply, and even particle size (43), likely affect denitrification (17, 73), only a few or even none would appear dominant in a complex environment where the multiple gradients may simultaneously increase and/or decrease DEA. For example, nitrate concentration, sometimes reported as an important or even primary factor affecting sediment denitrification (54, 63), may exert minimal control on DEA in marsh sediments (this study and reference 48) because of the coexisting salinity effect (48).
Besides elevation and carbon availability, selected metals were also found to be significantly related to community composition and to DEA. While some metals affected several microbial measurements (e.g., Cu for both profiles and cell counts and Ni for both profiles and DEA), others (e.g., V) were specific to denitrifier community composition. Although effects of metals on total microbial community composition have been widely studied (for reviews, see references 24 and 35), and similar metals were observed influencing 16S TRFLP profiles in two previous studies involving the CSM (12, 18), few assessments have been made for denitrification (47). Further, similar metal amendments caused site-specific responses for denitrification rate (47), suggesting that variations in the “transducers” (76) of denitrification, i.e., denitrifier communities, could modulate effects of metals on the denitrification process. In studies of denitrifying fractions of whole communities, using extracted cells from a sandy loam, significant reductions in denitrification rates and in denitrifier growth rates were caused by heavy metals (Cd, Cu, and Zn), even when communities were adapted to the metals (26). In the present study, Cu, Ni, V, Se, and Cr were present in the eluates at concentrations that caused potential adverse effects on denitrifiers (Table 1) and affected denitrifier community composition. Although the concentration range of Cu (0 to 277 ng g−1 dry sediment) (Table 1) was 1,000-fold lower than that recently reported for estuarine sediments (47) and 2 orders of magnitude lower than the dose used in the previous study (80 mg kg−1 dry soil), only a tiny fraction (0.70 to 0.95 mg kg−1) of the latter was extractable and thus bioavailable (26). The concentration range of Ni (0.03 to 0.3 mg l−1) was similar to a level reportedly negatively affecting denitrification rates and diminishing nirS genes to nondetectable levels in river biofilms (0.5 mg l−1) (42). In this study, however, the metal concentrations in eluates were measured and thus represented not only the bioavailable portion of the sediment metals but also the fraction affecting long-term exposure to microbes (12). Indeed, long-term exposures to microbes at metal concentrations in ranges comparable to those used in this study have been shown to be toxic when free metal ion concentrations were considered (24).
Indirectly, the observed similar gradients (elevation, DOC, and metals) of DEA and denitrifying community composition could imply that community structure and function are coupled. However, a lack of direct correlation between denitrifier activity and community composition indicates an uncoupling, as in a previous study (66). Yet, denitrifier community composition has been shown to affect denitrification rate (14, 30, 65) and its response to environmental conditions, such as oxygen level and pH (14). The apparent inconsistency may be explained in several ways. First, the presence of functional genes (e.g., nirS in the current study and nosZ in reference 66) does not ensure transcription and hence DEA (60). This might be referred to as “methodological uncoupling.” Second, since the ability to denitrify is widespread among diverse phylogenies, denitrifiers are functionally redundant (78), but meanwhile, enzymatic efficiency may vary greatly among species (22). Thus, shifts in community composition may not necessarily lead to changes in DEA of similar magnitudes. A plausible scenario would be that an environmental factor exerts effects on a highly abundant and/or efficient denitrifying group, and such effects are detected on both denitrifier community composition and DEA. Here, the heavy metal Ni (plus As and Rb) was significantly related to nirS TRFLP and also significantly affected DEA, which was similar to a prior report that Ni negatively impacted both nirS gene abundance and denitrification rate (42). Third, denitrifiers could be functionally complementary, as different subcommunities become active under different environmental conditions (60). For example, vanadium (V), although correlated with nirS community composition, did not correlate with DEA, perhaps because correlation between V and community composition may reflect the presence of vanadate-reducing denitrifiers (3, 4) causing a complementary shift (60) in community composition rather than inhibition of denitrification activity. Also, a positive correlation between V and DOC might have masked a correlation between V and DEA. Fourth, although both microbial community composition and environmental factors control denitrification activity (65), one or the other may dominate in different cases. Finally, although the cytochrome cd1-containing nitrite reductase (NirS) is generally more common (22), the copper-containing NirK protein may also be present and contribute to DEA in the current study. Perhaps Cu may induce a shift to NirK, thus influencing nirS TRFLP profiles while not significantly altering the overall DEA. Studies that include nirK TRFLP could be used to assess this possibility.
Acknowledgments
Funding for this work was provided by the U.S. Environmental Protection Agency's STAR Estuarine and Great Lakes (EaGLe) program through U.S. EPA agreement number R-882867601, by the University of California Marine Council's Coastal Environmental Quality Initiative through a graduate student fellowship, and by the University of California Natural Reserve System through a Mildred E. Mathias graduate student research grant, with support from the National Science Foundation under grant numbers OCE 9982105 and OCE 0620276.
We thank Tom Young for the availability of the inductively coupled plasma mass spectrometer and Scott Olson, Laurie Van De Werfhorst, and Daniel Brooks for their assistance. This work was performed at the University of California Natural Reserve System Carpinteria Salt Marsh Reserve.
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Aelion, C. M., and J. N. Shaw. 2000. Denitrification in South Carolina (USA) coastal plain aquatic sediments. J. Environ. Qual. 29:1696-1703. [Google Scholar]
- 2.Alongi, D. M. 1998. Coastal ecosystem processes. CRC Press, New York, NY.
- 3.Antipov, A. N., N. N. Lyalikova, T. V. Khijniak, and N. P. L'Vov. 2000. Vanadate reduction by molybdenum-free dissimilatory nitrate reductases from vanadate-reducing bacteria. IUBMB Life 50:39-42. [DOI] [PubMed] [Google Scholar]
- 4.Antipov, A. N., D. Y. Sorokin, N. P. L'Vov, and J. G. Kuenen. 2003. New enzyme belonging to the family of molybdenum-free nitrate reductases. Biochem. J. 369:185-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avrahami, S., R. Conrad, and G. Braker. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizer and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagwell, C. E., and C. R. Lovell. 2000. Persistence of selected Spartina alterniflora rhizoplane diazotrophs exposed to natural and manipulated environmental variability. Appl. Environ. Microbiol. 66:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blum, L. K., M. S. Roberts, J. L. Garland, and A. L. Mills. 2004. Distribution of microbial communities associated with the dominant high marsh plants and sediments of the United States east coast. Microb. Ecol. 48:375-388. [DOI] [PubMed] [Google Scholar]
- 8.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braker, G., J. Z. Zhou, L. Y. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burford, J. R., and J. M. Bremner. 1975. Relationships between denitrification capacities of soils and total, water-soluble, and readily decomposable soil organic matter. Soil Biol. Biochem. 7:389-394. [Google Scholar]
- 12.Cao, Y., G. N. Cherr, A. L. Córdova-Kreylos, T. W.-M. Fan, P. G. Green, R. M. Higashi, M. G. LaMontagne, K. M. Scow, C. A. Vines, J. Yuan, and P. A. Holden. 2006. Relationships between sediment microbial communities and pollutants in two California salt marshes. Microb. Ecol. 52:619-633. [DOI] [PubMed] [Google Scholar]
- 13.Carlson, C. A., S. J. Giovannoni, D. A. Hansell, S. J. Goldberg, R. Parsons, and K. Vergin. 2004. Interactions between DOC, microbial processes, and community structure in the mesopelagic zone of the northwestern Sargasso Sea. Limnol. Oceanogr. 49:1073-1083. [Google Scholar]
- 14.Cavigelli, M. A., and G. P. Robertson. 2000. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81:1402-1414. [Google Scholar]
- 15.Christian, R. R., K. Bancroft, and W. J. Wiebe. 1975. Distribution of microbial adenosine triphosphate in salt marsh sediments at Sapelo Island, Georgia. Soil Sci. 119:89-97. [Google Scholar]
- 16.Clarke, K. R., and R. M. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd edition. PRIMER-E, Ltd., Plymouth, United Kingdom.
- 17.Clement, J.-C., G. Pinay, and P. Marmonier. 2002. Seasonal dynamics of denitrification along topohydrosequences in three different riparian wetlands. J. Environ. Qual. 31:1025-1037. [DOI] [PubMed] [Google Scholar]
- 18.Córdova-Kreylos, A. L., Y. Cao, P. G. Green, H.-M. Hwang, K. Kuivila, M. G. LaMontagne, L. C. V. D. Werfhorst, P. A. Holden, and K. M. Scow. 2006. Diversity, composition, and geographical distribution of microbial communities in California salt marsh sediments. Appl. Environ. Microbiol. 72:3357-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis, J. L., B. L. Nowicki, and C. Wigand. 2004. Denitrification in fringing salt marshes of Narragansett Bay, Rhode Island, USA. Wetlands 24:870-878. [Google Scholar]
- 20.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engebretson, J. J., and C. L. Moyer. 2003. Fidelity of select restriction endonucleases in determining microbial diversity by terminal-restriction fragment length polymorphism. Appl. Environ. Microbiol. 69:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firestone, M. K. 1982. Biological denitrification, p. 289-326. In F. J. Stevenson (ed.), Nitrogen in agriculture soil. American Society of Agronomy, Inc., Crop Science Society of America, Inc., and Soil Science Society of America, Inc., Madison, WI.
- 23.Gardner, W. H. 1986. Water content, p. 504-507. In A. Klute (ed.), Methods of soil analysis, part 1: physical and mineralogical methods, 2nd ed. American Society of Agronomy, Inc., and Soil Science Society of America, Inc., Madison, WI.
- 24.Giller, K. E., E. Witter, and S. P. McGrath. 1998. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol. Biochem. 30:1389-1414. [Google Scholar]
- 25.Holden, P. A., M. G. LaMontagne, A. K. Bruce, W. G. Miller, and S. E. Lindow. 2002. Assessing the role of Pseudomonas aeruginosa surface-active gene expression in hexadecane biodegradation in sand. Appl. Environ. Microbiol. 68:2509-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtan-Hartwig, L., M. Bechmann, T. Risnes Hoyas, R. Linjordet, and L. Reier Bakken. 2002. Heavy metals tolerance of soil denitrifying communities: N2O dynamics. Soil Biol. Biochem. 34:1181-1190. [Google Scholar]
- 27.Holtan-Hartwig, L., P. Dorsch, and L. R. Bakken. 2000. Comparison of denitrifying communities in organic soils: kinetics of NO3− and N2O reduction. Soil Biol. Biochem. 32:833-843. [Google Scholar]
- 28.Howes, B. L., P. K. Weiskel, D. D. Geohringer, and J. M. Teal. 1996. Interception of freshwater and nitrogen transport from uplands to coastal waters: the role of salt marshes, p. 287-310. In K. F. Nordstrom and C. T. Roman (ed.), Estuarine shores: evolution, environments and human alterations. John Wiley & sons, New York, NY.
- 29.Hunt, P. G., T. A. Matheny, and K. C. Stone. 2004. Denitrification in a coastal plain riparian zone contiguous to a heavily loaded swine wastewater spray field. J. Environ. Qual. 33:2367-2374. [DOI] [PubMed] [Google Scholar]
- 30.Jayakumar, D. A., C. A. Francis, S. W. A. Naqvi, and B. B. Ward. 2004. Diversity of nitrite reductase gene (nirS) in the denitrifying water column of the coastal Arabian Sea. Aquat. Microb. Ecol. 34:69-78. [Google Scholar]
- 31.Keith-Roach, M. J., N. D. Bryan, R. D. Bardgett, and F. R. Livens. 2002. Seasonal changes in the microbial community of a salt marsh, measured by phospholipid fatty acid analysis. Biogeochemistry 60:77-96. [Google Scholar]
- 32.Kendall, M. G. 1970. Rank correlation methods, 4th ed. Griffin, London, United Kingdom.
- 33.Knowles, R. 1982. Denitrification. Microbiol. Rev. 46:43-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch, M. S., E. Maltby, G. A. Oliver, and S. A. Bakker. 1992. Factors controlling denitrification rates of tidal mudflats and fringing salt marshes in south-west England. Estuarine Coast Shelf Sci. 34:471-485. [Google Scholar]
- 35.Kozdroj, J., and J. D. van Elsas. 2001. Structural diversity of microorganisms in chemically perturbed soil assessed by molecular and cytochemical approaches. J. Microbiol. Methods 43:197-212. [DOI] [PubMed] [Google Scholar]
- 36.Kutner, M. H., C. J. Nachtsheim, and J. Neter. 2004. Applied linear regression models, 4th ed. McGraw-Hill, Irvine, CA.
- 37.Lachat Instruments. 1995. QuikChem method 10-107-06-5-A. In Methods list for automated ion analyzers. Lachat Instruments, Milwaukee, WI.
- 38.Lachat Instruments. 1996. QuikChem method 10-107-04-1-C. In Methods list for automated ion analyzers. Lachat Instruments, Milwaukee, WI.
- 39.Lachat Instruments. 1996. QuikChem method 31-115-01-3-A. In Methods list for automated ion analyzers. Lachat Instruments, Milwaukee, WI.
- 40.LaMontagne, M. G., I. Leifer, S. Bergmann, L. C. Van De Werfhorst, and P. A. Holden. 2004. Bacterial diversity in marine hydrocarbon seep sediments. Environ. Microbiol. 6:799-808. [DOI] [PubMed] [Google Scholar]
- 41.Law, C. S., A. P. Rees, and J. P. Owens. 1991. Temporal variability of denitrification in estuarine sediments. Estuarine Coast Shelf Sci. 33:37-56. [Google Scholar]
- 42.Lawrence, J. R., M. R. Chenier, R. Roy, D. Beaumier, N. Fortin, G. D. W. Swerhone, T. R. Neu, and C. W. Greer. 2004. Microscale and molecular assessment of impacts of nickel, nutrients, and oxygen level on structure and function of river biofilm communities. Appl. Environ. Microbiol. 70:4326-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lensi, R., A. Clays-Josserand, and L. J. Monrozier. 1995. Denitrifiers and denitrifying activity in size fractions of a mollisol under permanent pasture and continuous cultivation. Soil Biol. Biochem. 27:61-69. [Google Scholar]
- 44.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu, X., S. M. Tiquia, G. Holguin, L. Wu, S. C. Nold, A. H. Devol, K. Luo, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity of denitrifying genes in continental margin sediments within the oxygen deficient zone of the Pacific Coast of Mexico. Appl. Environ. Microbiol. 69:3549-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovell, C. R., C. E. Bagwell, M. Czako, L. Marton, Y. M. Piceno, and D. B. Ringelberg. 2001. Stability of a rhizosphere microbial community exposed to natural and manipulated environmental variability. FEMS Microbiol. Ecol. 38:69-76. [Google Scholar]
- 47.Magalhães, C. M., J. Costa, C. Teixeira, and A. A. Bordalo. 2007. Impact of trace metals on denitrification in estuarine sediments of the Douro River estuary, Portugal. Mar. Chem. 107:332-341. [Google Scholar]
- 48.Magalhães, C. M., S. B. Joye, R. M. Moreira, W. J. Wiebe, and A. A. Bordalo. 2005. Effect of salinity and inorganic nitrogen concentrations on nitrification and denitrification rates in intertidal sediments and rocky biofilms of the Douro River estuary, Portugal. Water Res. 39:1783-1794. [DOI] [PubMed] [Google Scholar]
- 49.Mergel, A., O. Schmitz, T. Mallmann, and H. Bothe. 2001. Relative abundance of denitrifying and dinitrogen-fixing bacteria in layers of a forest soil. FEMS Microbiol. Ecol. 36:33-42. [DOI] [PubMed] [Google Scholar]
- 50.Montague, C. L., and H. T. Odum. 1997. Setting and functions, p. 9-35. In C. L. Coultas and Y.-P. Hsieh (ed.), Ecology and management of tidal marshes: a model from the Gulf of Mexico. St. Lucie Press, Delray Beach, FL.
- 51.Mounier, E., S. Hallet, D. Chèneby, E. Benizri, Y. Gruet, C. Nguyen, S. Piutti, C. Robin, S. Slezack-Deschaumes, F. Martin-Laurent, J. C. Germon, and L. Philippot. 2004. Influence of maize mucilage on the diversity and activity of the denitrifying community. Environ. Microbiol. 6:301-312. [DOI] [PubMed] [Google Scholar]
- 52.Nelson, D. W., and L. E. Sommers. 1996. Total carbon, organic carbon, and organic matter, p. 1002-1005. In D. L. Sparks (ed.), Methods of soil analysis, part 3: chemical methods. Soil Science Society of America, Inc., and American Society of Agronomy, Inc., Madison, WI.
- 53.Nogales, B., K. N. Timmis, D. B. Nedwell, and A. M. Osborn. 2002. Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowicki, B. L. 1994. The effect of temperature, oxygen, salinity, and nutrient enrichment on estuarine denitrification rates measured with a modified nitrogen gas flux technique. Estuarine Coast Shelf Sci. 38:137-156. [Google Scholar]
- 55.Nowicki, B. L., E. Requintina, D. Van Keuren, and J. Portnoy. 1999. The role of sediment denitrification in reducing groundwater-derived nitrate inputs to Nauset Marsh Estuary, Cape Cod, Massachusetts. Estuaries 22:245-259. [Google Scholar]
- 56.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Page, H. M., R. L. Petty, and D. E. Meade. 1995. Influence of watershed runoff on nutrient dynamics in a Southern California salt marsh. Estuarine Coast Shelf Sci. 41:163-180. [Google Scholar]
- 58.Palmer, M. W. 1993. Putting things in even better order: the advantages of canonical correspondence analysis. Ecology 74:2215-2230. [Google Scholar]
- 59.Parkin, T. B. 1987. Soil microsites as a source of denitrification variability. Soil Sci. Soc. Am. J. 51:1194-1199. [Google Scholar]
- 60.Philippot, L., and S. Hallin. 2005. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr. Opin. Microbiol. 8:234-239. [DOI] [PubMed] [Google Scholar]
- 61.Piceno, Y. M., and C. R. Lovell. 2000. Stability in natural bacterial communities: I. Nutrient addition effects on rhizosphere diazotroph assemblage composition. Microb. Ecol. 39:32-40. [DOI] [PubMed] [Google Scholar]
- 62.Piceno, Y. M., and C. R. Lovell. 2000. Stability in natural bacterial communities: II. Plant resource allocation effects on rhizosphere diazotroph assemblage composition. Microb. Ecol. 39:41-48. [DOI] [PubMed] [Google Scholar]
- 63.Poe, A. C., M. F. Piehler, S. P. Thompson, and H. W. Paerl. 2003. Denitrification in a constructed wetland receiving agricultural runoff. Wetlands 23:817-826. [Google Scholar]
- 64.Priemé, A., G. Braker, and J. M. Tiedje. 2002. Diversity of nitrite reductase (nirK and nirS) gene fragments in forested upland and wetland soils. Appl. Environ. Microbiol. 68:1893-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rich, J. J., R. S. Heichen, P. J. Bottomley, K. J. Cromack, and D. D. Myrold. 2003. Community composition and functioning of denitrifying bacteria from adjacent meadow and forest soils. Appl. Environ. Microbiol. 69:5974-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rich, J. J., and D. D. Myrold. 2004. Community composition and activities of denitrifying bacteria from adjacent agricultural soil, riparian soil, and creek sediment in Oregon, USA. Soil Biol. Biochem. 36:1431. [Google Scholar]
- 67.Sanger, D. M., A. F. Holland, and G. I. Scott. 1999. Tidal creek and salt marsh sediments in South Carolina coastal estuaries: I. Distribution of trace metals. Arch. Environ. Contam. Toxicol. 37:445. [DOI] [PubMed] [Google Scholar]
- 68.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.ter Braak, C. J. F. 1987. The analysis of vegetation-environmental relationships by canonical correspondence analysis. Vegetatio 69:69-77. [Google Scholar]
- 70.ter Braak, C. J. F., and P. Smilauer. 2002. CANOCO reference manual and CanoDraw for Windows user's guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY.
- 71.Thompson, S. P., H. W. Paerl, and M. Go. 1995. Seasonal patterns of nitrification and denitrification in a natural and a restored salt marsh. Estuaries 18:399-408. [Google Scholar]
- 72.Tiedje, J. M. 1994. Denitrifiers, p. 245-267. In R. W. Weaver, S. J. Angle, P. J. Bottomley, D. F. Bezdicek, S. Smith, M. A. Tabatabai, and A. G. I. Wollum (ed.), Method of soil analysis, part 2: microbiological and biochemical properties, vol. 5. Soil Science Society of America, Inc., Madison, WI. [Google Scholar]
- 73.Tomaszek, J. A., and E. Czerwieniec. 2003. Denitrification and oxygen consumption in bottom sediments: factors influencing rates of processes. Hydrobiologia 504:59-65. [Google Scholar]
- 74.U.S. Environmental Protection Agency. 1994. Method 6020. In Test methods for evaluating solid waste: physical/chemical methods. Publication SW-846. EPA Office of Solid Waste, Washington, DC.
- 75.Valiela, I., and M. L. Cole. 2002. Comparative evidence that salt marshes and mangroves may protect seagrass meadows from land-derived nitrogen loads. Ecosystems 5:92-102. [Google Scholar]
- 76.Wallenstein, M. D., D. D. Myrold, M. Firestone, and M. Voytek. 2006. Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16:2143-2152. [DOI] [PubMed] [Google Scholar]
- 77.Wolsing, M., and A. Prieme. 2004. Observation of high seasonal variation in community structure of denitrifying bacteria in arable soil receiving artificial fertilizer and cattle manure by determining T-RFLP of nir gene fragments. FEMS Microbiol. Ecol. 48:261-271. [DOI] [PubMed] [Google Scholar]
- 78.Zumft, W. G. 1992. The denitrifying prokaryotes, p. 443-582. In A. Balows, H. G. Truper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer, Heidelberg, Germany.