Abstract
Altered expression of the human FEV (fifth Ewing variant) ETS transcription factor gene impacts the level of CNS serotonin (5-HT) neuron gene expression and maternal nurturing. However, the regulatory mechanisms that determine FEV expression are poorly understood. Here, we investigated the cis-regulatory control of FEV to begin to identify the upstream transcription factors that restrict FEV expression to 5-HT neurons. We find that sequences extending only 275 bp upstream of the FEV 5′ untranslated region are sufficient to direct FEV transgene expression to embryonic 5-HT neurons, although sequences farther upstream are required for maintenance in adult 5-HT neurons. Two highly conserved consensus GATA factor binding sites within the 275 bp region interact with GATA factors in vitro. Chromatin immunoprecipitations with embryonic hindbrain demonstrated Gata-2 interactions with the orthologous mouse Pet-1 ETS cis-regulatory region. Mutagenesis of GATA sites revealed that one or the other site is required for serotonergic FEV transgene expression. Unexpectedly, FEV–LacZ transgenes enabled determination of 5-HT neuron precursor fate in the adult Pet-1−/− dorsal and median raphe nuclei and thus provided additional insight into FEV/Pet-1 function. Comparable numbers of FEV–LacZ-positive cells were detected in Pet-1+/− and Pet-1−/− adult dorsal raphe nuclei, indicating that the majority of mutant serotonergic precursors are not fated to apoptosis. However, B7 dorsal raphe cells were aberrantly distributed, suggesting a role for FEV/Pet-1 in their midline organization. Our findings identify a direct transcriptional interaction between Gata-2 and FEV and a unique marker for new insight into FEV/Pet-1 function in 5-HT neuron development.
Keywords: FEV, Pet-1, GATA-2, serotonin, cis-regulatory, transgenic
Introduction
Alterations in serotonin (5-HT) neuron gene expression lead to changes in emotional behavior in rodents (Ansorge et al., 2007; Beaulieu et al., 2008) and are associated with depression and anxiety disorders in humans (Ansorge et al., 2007; Murphy and Lesch, 2008). A critical developmental determinant of serotonergic gene expression is the ETS transcription factor Pet-1 (Hendricks et al., 2003). In Pet-1−/− mice, normal numbers of serotonergic precursors are generated in the ventral hindbrain, but most fail to express tryptophan hydroxylase 2 (TPH2), the serotonin transporter (SERT), and other serotonergic genes. The dramatically reduced levels of 5-HT in Pet-1−/−-deficient mice is associated with delayed respiratory maturation (Erickson et al., 2007), increased aggression and anxiety-like behavior in adulthood (Hendricks et al., 2003), and a profound deficit in maternal behavior (Lerch-Haner et al., 2008).
Pet-1 expression is governed by a serotonergic transcriptional cascade that includes the proneural factor Mash1 (Pattyn et al., 2004), the homeodomain factor Nkx2.2 (Pattyn et al., 2003), and the forkhead box factor Foxa2 (Jacob et al., 2007) in ventral hindbrain progenitors and the zinc finger factor Gata-2 in postmitotic precursors (Craven et al., 2004). We showed previously that a cis-regulatory region upstream of Pet-1 is sufficient to direct transgene reporter expression to developing and adult 5-HT neurons (Scott et al., 2005). Therefore, this region is a target of upstream serotonergic transcriptional cascade. However, the precise location of cis-regulatory elements for serotonergic expression of Pet-1 has not been determined, nor is it known whether any of the identified transcription factors in the cascade directly regulate Pet-1.
Human FEV encodes a protein that has 96% identity to Pet-1 and is expressed specifically in human raphe (Iyo et al., 2005). Recently, we showed that both serotonergic and nurturing deficits in Pet-1−/− mice could be rescued with a bacterial artificial chromosome encoding FEV (Lerch-Haner et al., 2008), hence demonstrating that FEV is an ortholog of Pet-1. Importantly, the levels of serotonergic gene expression in rescued Pet-1−/− mice, as well as maternal care and offspring survival, depended on the level of FEV gene expression. These findings show subtle alterations in FEV expression can influence serotonergic gene expression and the quality of nurturing behaviors. Thus, FEV regulation and function may be relevant to disease pathogenesis (Rand et al., 2007). However, the mechanisms that control FEV expression in 5-HT neurons have not been investigated.
Here, we investigated the cis-regulatory control of FEV and report that sequences surrounding the FEV transcriptional start site are sufficient to direct 5-HT neuron-specific transgene expression. Two conserved GATA sites in this region are required in a functionally redundant manner for serotonin neuron transgene expression. Finally, FEV-directed transgenes enable fate analysis of mutant 5-HT neuron precursors in the adult Pet-1−/− brain and show that these mutant precursors are maintained but are aberrantly distributed. Our findings provide insight into the regulation and function of conserved human and mouse serotonergic developmental control genes and a transcriptional control map for future identification of functional FEV cis-regulatory variants.
Materials and Methods
Transgenes
FEV2.2Z, FEV1.1Z, and FEV0.6Z.
A BGZA vector (Yee and Rigby, 1993) was first modified to introduce BglII and PacI sites in the multiple cloning site, and then a 2.2 kb BglII/PacI FEV upstream fragment was subcloned into the modified BGZA vector. The vector sequences were removed before pronuclear injection with EagI. FEV1.1Z and FEV0.6Z were prepared by KpnI/EagI or StuI/EagI digests, respectively, of the FEV2.2Z plasmid.
FEV2.2ΔZ and FEV1.1ΔZ.
PCR was used to amplify products on either side of the deletion (Fig. 1) using the following primers: 5′-GGGCAGATCTGAATCTGCCCTTGACTGAGC-3′ and 5′-TAATGCGCGCCTTCCCCGTCTCCTTCTTG-3′; 5′-TAATGCGCGCGAAGAAGGGCTCTAGGGAGG-3′ and 5′-GGCGTCTAGACCTTAATTAAAGCGCTGCCG-3′ (underlining indicates BglII, BssHII, or PacI sites used to clone in the amplified products). PCR products were inserted into the corresponding BglII, BssHII, or PacI sites in the FEV2.2Z plasmid and sequenced. Relative to the predicted start site, sequences −214/+25 were removed. FEV2.2ΔZ and FEV1.1ΔZ were digested with EagI or KpnI/EagI before injection.
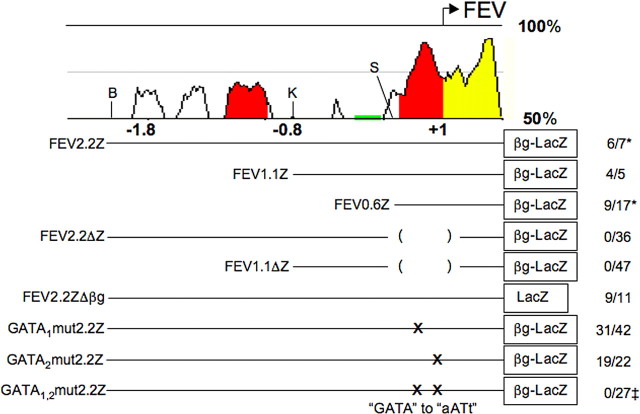
Figure 1.
Conserved FEV/Pet-1 upstream sequences and transgene structure. Top, zPicture analysis of mouse and human conserved genomic sequences upstream of FEV reveals blocks of human/mouse conservation. The y-axis shows percentage sequence identity. Colored peaks indicate significant sequence conservation, defined as at least 70% identity over 100 bp. Red peaks indicate predicted intergenic sequences, yellow indicates untranslated exons, and green indicates repetitive sequences. Restriction sites used to prepare transgenes are indicated. B, BglII; K, KpnI; S, StuI. Numbering is relative to the transcriptional start site as in the NCBI annotation. Bottom, Structure of FEV LacZ transgenes tested in this study. The 5′ ends of FEV2.2Z, FEV1.1Z, and FEV0.6Z are located at −1924, −787, and −275 bp, respectively, relative to the transcriptional start site. The 3′ end of all transgenes is a PacI site at nucleotide +318 in the 5′ untranslated exon. βg, β-Globin TATA site. Right, Number of transgenic lines with expression patterns similar to endogenous Pet-1 at E12.5, over the total number of lines evaluated for each construct. ‡, Very weak expression detected in 11 of 27 lines. *Adult expression also examined: 11 of 12 FEV2.2Z and 1 of 4 FEV0.6Z lines showed adult serotonergic transgene expression.
FEV2.2ZΔβg.
FEV2.2Z was digested with PacI and SbfI to excise the β-globin TATA box, and overhanging ends were blunted and self-ligated to generate FEV2.2ZΔβg. Reporters were verified by sequencing before pronuclear injection. The vector backbone sequences were removed by EagI digest.
GATA mutants.
For mutagenesis of GATA binding sites, the QuikChange Site-Directed Mutagenesis kit (Stratagene) was used. Following a previously described strategy (Kobayashi-Osaki et al., 2005), the GATA sites were destroyed either singly or in combination by modification of the “GATA” motif to “AATT” using the following mutagenic primers (underlined residues were modified): FEV distal site (GATA1) 5′-GGATGCGGGCAGAGATAAAGGGAGCAACGGCTGC-3′ and complement; FEV proximal site (GATA2) 5′-GGAAATTTAAAAGTGAAGATGCAGATAACGCAGCCTGGAGACGGG-3′ and complement. The inserts were fully sequenced, and EagI digest was used to release the inserts before pronuclear injection.
FEV60Z.
Genomic DNA was obtained from a human bacterial artificial chromosome RPCI-3304 (GenBank accession number AC097468). A pBACe3.6 vector (Frengen et al., 1999) was modified by the insertion of a polylinker containing PmeI and RsrII restriction sites into the MluI site (Scott et al., 2005). A 58.6 kb NruI/RsrII FEV fragment was obtained from RPCI-3304 and subcloned into pBACe3.6 using PmeI and RsrII sites. The β-globin TATA, lacZ, and simian virus 40 polyadenylation sequences were released from a BGZA plasmid (Yee and Rigby, 1993) with an EagI/NotI digest and inserted into the NotI site of pBACe3.6 downstream of the FEV fragment to prepare FEV60Z. The vector backbone of FEV60Z transgene was removed with AscI before pronuclear injection.
Transgenic mice
Pronuclear injections were performed by the Case Transgenic and Targeting Facility as described previously (Scott et al., 2005). FEV60Z founders were identified by PCR with 5′-CAAAGACAGGAGGAGGTTGGTAGC-3′ and 5′-TTGGGTAACGCCAGGGTTTTCC-3′ primers. All other transgenes were identified with 5′-CTTTATTGCGATGGGACGAT-3′ and 5′-GTTTTCCCAGTCACGACGTT-3′. Cycling conditions were as follows: 95°C for 5 min; 35 cycles of 30 s at 94°C, 30 s at 60°C, then 60 s at 72°C; then 72°C for 7 min. Products were 260 bp. Pet-1−/− and Pet-1+/− mice were generated and genotyped as described previously (Hendricks et al., 2003). Twelve lines were generated for FEV2.2Z, seven of which were analyzed at embryonic day 12.5 (E12.5). For FEV0.6Z, 17 founders/lines were analyzed at E12.5. Three additional FEV0.6Z lines were generated but were excluded from our analyses because of a high level of ectopic hindbrain transgene expression. For FEV1.1Z, FEV2.2ΔZ, FEV1.1ΔZ, FEV2.2ZΔβg, and GATA mutant lines, founder E12.5 embryos were evaluated exclusively by whole-mount 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) staining, followed in certain lines by staining for 5-HT.
Histology
Adult mice were transcardially perfused for 15–20 min with ice-cold 4% paraformaldehyde (PFA) in 0.1 m PBS, postfixed for 4 h at 4°C, and then sunk overnight in 20% sucrose in 0.1 m PBS. Embryos were fixed by immersion in 4% PFA in 0.1 m PBS for 10 min (whole-mount X-gal staining) or 1 h (slices). X-gal staining was performed on fixed whole embryos as described previously (Scott et al., 2005); otherwise, embryos were sunk in 30% sucrose overnight at 4°C, then frozen in Tissue Freezing Medium, and stored at −80°C. Tissue sections were obtained using the freezing microtome (adult, 20 μm) or cryostat (embryo, 10–20 μm). In some cases, after whole-mount X-gal staining, embryos were fixed, sliced, and stained for 5-HT as described previously (Hendricks et al., 1999). Fluorescent immunohistochemistry for β-galactosidase (β-gal), 5-HT, TPH, and neuronal-specific nuclear protein (NeuN) was performed as described previously (Scott et al., 2005). Gata-2 immunostaining was performed with RC1.1 rat anti-chicken Gata-2 antibody (gift from J. D. Engel, University of Michigan, Ann Arbor MI). Other antisera used were mouse anti-Gata-3 (Santa Cruz Biotechnology), chicken anti-tyrosine hydroxylase (TH) (Aves), and goat anti-choline acetyltransferase (ChAT) (Millipore Bioscience Research Reagents). Gata-3 immunostaining required a mild antigen retrieval treatment (30 min incubation at 37°C in 10 mm citric acid, pH 3) before incubation with primary antibody.
Sequence analysis
Human and mouse sequence comparisons were performed as described previously (Scott et al., 2005). The minimal criteria for significant sequence conservation were 70% identity over 100 bp. Gene annotation information was derived from NCBI (FEV GenBank accession number NM_017521; Pet-1 GenBank accession number NM_153111) and the ECR browser tools (Ovcharenko et al., 2004). Predicted transcription factor binding sites were obtained using rVista 2.0 (Loots and Ovcharenko, 2004) and MatInspector (Cartharius et al., 2005). FEV-proximal conserved sequences were identified using a discontiguous megaBLAST search of the Whole Genome Survey database and Trace Archives [National Center for Biotechnology Information (NCBI)] and aligned using CLUSTALW (MacVector 7).
Electrophoretic mobility shift assays
Assays were performed with the LightShift EMSA kit (Pierce) using the supplied binding buffer supplemented with 1 mm EDTA (except for experiments with recombinant protein or antibody). GATA sites upstream of FEV were tested with the following biotinylated oligonucleotides (GATA motif underlined): GATA1 site, 5′-CGGGCAGAGATAAAGGGAGC-3′; GATA2 site, 5′-AAGATGCAGATAACGCAGCC-3′; and complementary oligonucleotides. Biotin-labeled oligonucleotides were annealed, and 60–80 fmol of double-stranded oligonucleotides were incubated with ∼1 μg recombinant Gata-1 protein (Panomics) or 6.4–12.8 μg of HeLa nuclear extracts (Promega). Competition assays were performed using 100-fold excess of unlabeled wild-type or base-substituted oligonucleotides (in which the GATA motif was changed to AATT as in transgenic studies). For supershift experiments, 5 μl of goat anti-Gata-2 (Santa Cruz Biotechnology) or rabbit anti-green fluorescent protein (GFP) (Invitrogen) were used. For both supershift and competition experiments, extracts were preincubated for 20 min in the absence of labeled DNA, followed by 20 min incubation with labeled oligonucleotide. Reactions were electrophoresed on 6% PAGE in 0.5× Tris-borate-EDTA and processed according the instructions of the manufacturer (Pierce).
Chromatin immunoprecipitation assays
Hindbrain tissues (from mesencephalic flexure to cervical flexure) were removed from 141 E11.5 embryos and quick frozen on dry ice. Gata-2 occupancy of genomic regions was tested by GenPathway, using rabbit anti-Gata-2 antibody (Santa Cruz Biotechnology) and quantitative PCR (QPCR) according to their protocols (Alexiadis et al., 2007). Supplemental Table 1 (available at www.jneurosci.org as supplemental material) gives the sequences of primers used for QPCR. Each primer pair gave a single product by melt-curve analysis and agarose gel electrophoresis. Binding was tested in triplicate for two negative control regions (untranscribed genomic regions Untr8, Untr17) and several FEV upstream regions containing predicted GATA sites. Data are expressed as fold increase in binding for each sample relative to binding at Untr17. Differences in binding among regions were calculated using one-way ANOVA with Bonferroni's multiple comparison test (Prism 4.0). Replication of the entire assay gave similar results.
Cell counts
A template for counting was generated in Adobe Photoshop CS3 (Adobe Systems) and consisted of four identical boxes centered on an image of a wild-type section outside the main body of the B7 dorsal raphe nucleus (DRN) (see Fig. 8E). Images of knock-out and control B7 were overlaid with this same template, centered at the midline using the ventricle and midline nuclei, and all cells within these boxes were counted. The mean number of cells within the boxes per section was averaged for each animal, and groups (knock-out or control) were compared using a two-tailed t test. A total of eight Pet-1−/− and six control (five Pet-1+/− and one Pet-1+/+) adult animals were counted, three to seven sections (mean of 5) per animal.
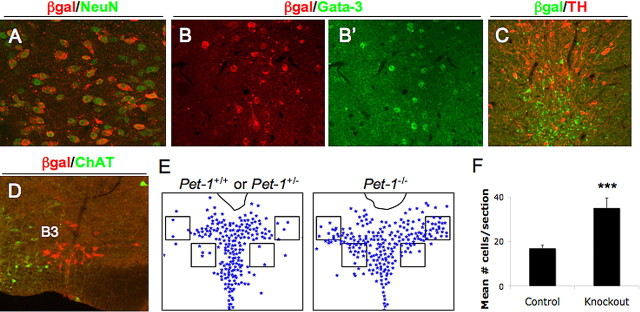
Figure 8.
Pet-1−/− cells in the B7 DRN retain serotonergic character but are aberrantly distributed. A–C, Coimmunostaining of FEV60Z, Pet-1−/− DRN for β-gal and NeuN (A), β-gal (B) and Gata-3 (B′), and β-gal and TH (C). D, Coimmunostaining of Pet-1−/− B3 ventrolateral medulla for β-gal and ChAT. E, Schematic of control and knock-out sections overlaid with the counting template. F, Mean + SEM number of cells per section in boxes in control (16.7 ± 1.5; n = 5 Pet-1+/−; n = 1 Pet-1+/+) and Pet-1−/− (34.9 ± 4; n = 8) brains at the level of B7 dorsal raphe. ***p = 0.003, two-tailed t test.
Results
Human FEV cis-regulatory elements direct expression to developing and adult 5-HT neurons
Rodent 5-HT neurons develop in two ventral hindbrain domains, a rostral domain extending from rhombomere (r) r1 to r3, and a caudal domain extending from r5 to r7 (Lidov and Molliver, 1982; Wallace and Lauder, 1983; Hendricks et al., 1999). Pet-1 expression is initiated at E10.5–E11 in the mouse rostral domain, followed ∼1 d later by expression in the caudal domain (Pfaar et al., 2002; Pattyn et al., 2003). In the adult brain, a large number of 5-HT neurons are present in the midline raphe nuclei, although significant numbers are also found scattered in more lateral regions of the pontine reticular formation and ventrolateral medulla (Steinbusch, 1981). Expression of the endogenous Pet-1 gene is maintained in what appears to be all 5-HT neurons of the adult brain (Hendricks et al., 1999).
Our previous studies showed that a 1.8 kb fragment immediately upstream of mouse Pet-1 was sufficient to direct transgene expression in embryonic and adult 5-HT neurons (Scott et al., 2005). Comparison of the Pet-1 and FEV upstream regions revealed two blocks of sequence conservation (Fig. 1). The proximal block includes the FEV 5′ untranslated exon and extends 310 bp into the FEV upstream region. The distal block comprises three subregions, the first of which begins 895 bp upstream of the FEV start site. To determine whether or not 5-HT neuron-specific elements are located among these sequences, we isolated a 2.2 kb FEV genomic fragment whose 3′ end is located in the FEV 5′ untranslated region (Fig. 1). This fragment was ligated upstream of a β-globin TATA box/lacZ/poly(A) signal sequence cassette (Yee and Rigby, 1993; Helms et al., 2000) to prepare transgene reporter FEV2.2Z. The β-globin TATA box was included in this and other transgenes to ensure the presence of a start site because neither the Pet-1 nor FEV promoters have been experimentally localized.
Seven FEV2.2Z lines were analyzed for expression at embryonic stages. Similar to endogenous Pet-1, FEV2.2Z expression was initiated in the rostral hindbrain between E10.5 and E11 (data not shown) and then expanded into the caudal domain by E12.5 (Fig. 2A) in six of seven lines (Fig. 1). Coimmunostaining with anti-5-HT and anti-β-gal revealed a 5-HT neuron-specific pattern of FEV2.2Z expression in the hindbrain (Fig. 2B). Expression of FEV2.2Z, however, was weak in the posterior half of the caudal hindbrain near the cervical flexure (Fig. 2C,C′) in all but one line.
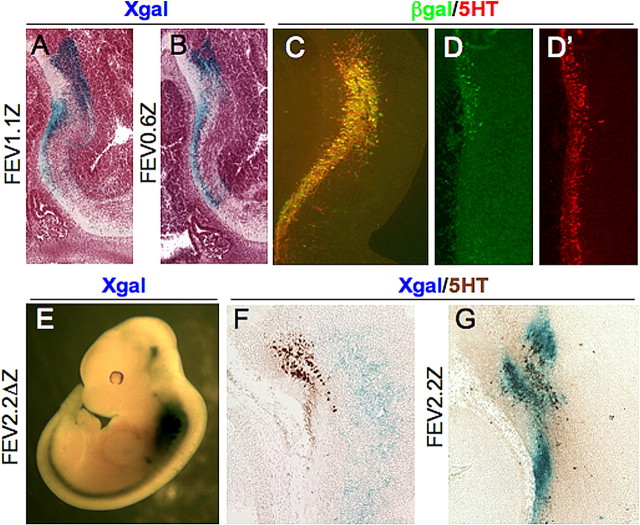
Figure 2.
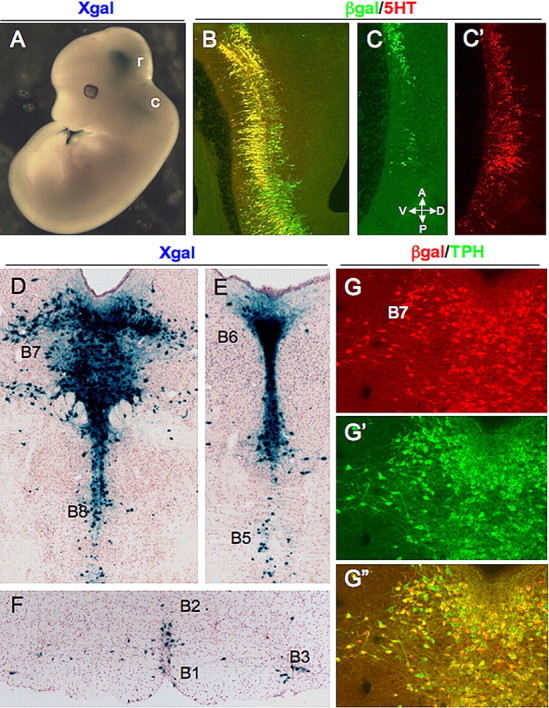
FEV is controlled by a cis-regulatory region that directs expression to developing and adult 5-HT neurons. A, Whole-mount X-gal staining of an E12.5 FEV2.2Z embryo. B–C′, Sagittal views of 5-HT and β-gal colocalization in the rostral (B) and caudal (C, C′) hindbrain. D–F, X-gal staining of adult coronal sections showing LacZ expression in the B7 DRN and B8 MRN (D), pontine dorsal (B6) and median (B5) raphe (E), and raphe obscurus (B2), raphe pallidus (B1), and ventrolateral medulla (B3) (F). G–G″, Immunostaining of the lateral wing region of the dorsal raphe B7 nucleus. G, Anti-β-gal; G′, anti-TPH; G″, merge of G and G′; r, rostral 5-HT neurons; c, caudal 5-HT neurons; A, anterior; D, dorsal; P posterior; V, ventral.
In 11 of 12 lines, adult FEV2.2Z expression was reproducibly present in the DRN and median raphe nuclei (MRN) as well as in the reticular formation and ventrolateral medulla (Fig. 2D–F and data not shown). Coimmunostaining of FEV2.2Z mice indicated that the majority of β-gal-positive (β-gal+) cells expressed TPH, the rate-limiting enzyme for 5-HT biosynthesis, in each of the adult raphe nuclei (Fig. 2G–G″). However, a small number of β-gal-negative (β-gal−)/TPH+ cells was observed in all FEV2.2Z lines (Fig. 2G″), and FEV2.2Z expression was missing from most 5-HT neurons in the B2 raphe obscurus (data not shown) in all but one line. These cells likely correspond to cells derived from the posterior half of the embryonic caudal hindbrain, and therefore additional cis-regulatory elements probably direct expression to this subdomain. Nevertheless, these findings show that a conserved regulatory region upstream of FEV directs transgene expression to developing and adult 5-HT neurons.
FEV promoter sequences are necessary and sufficient for embryonic serotonergic transgene expression
We next tested several additional FEV transgenes to determine the location of essential serotonergic cis-regulatory elements within the FEV 2.2 kb fragment. To investigate the importance of distal conserved sequences, we generated two transgenes with 5′ truncations of either 1.1 or 1.6 kb (FEV1.1Z or FEV0.6Z, respectively) (Fig. 1). Similar to FEV2.2Z, both FEV1.1Z and FEV0.6Z expression was restricted to the rostral and caudal hindbrain (Fig. 3A,B). Additional analysis of FEV0.6Z indicated that FEV0.6Z was expressed in a 5-HT neuron-specific pattern in the hindbrain (Fig. 3C), but expression was again lacking in posterior caudal 5-HT neurons (Fig. 3D,D′). Thus, important cis-regulatory elements for embryonic expression are not likely to be associated with the distal conserved sequences. These findings additionally suggest that conserved sequences surrounding the FEV transcriptional start site (−275/+318) constitute a minimal FEV cis-regulatory region sufficient for 5-HT neuron-specific transgene expression in the developing hindbrain. However, when four FEV0.6Z lines showing restricted serotonergic expression of LacZ in the embryonic hindbrain were evaluated at adult stages, only one of them continued to express LacZ, and this expression was very weak (data not shown). Thus, additional regulatory elements that are present in FEV2.2Z but missing in FEV0.6Z are likely to be required for maintenance of expression.
Figure 3.
Proximal conserved sequences are necessary and sufficient for 5-HT neuron-specific expression in the developing hindbrain. A, Sagittal view of E12.5 FEV1.1Z expression in the rostral hindbrain after whole-mount X-gal staining. B, C, Sagittal sections of E12.5 FEV0.6Z embryos after whole-mount X-gal staining (B) or 5-HT and β-gal coimmunostaining (C) in the rostral hindbrain. D, D′, Sagittal view of β-gal (D) and 5-HT immunostaining (D′) of caudal hindbrain from an E12.5 FEV0.6Z embryo. E, Whole mount X-gal staining of an E12.5 FEV2.2ΔZ founder. F, G, Sagittal views of rostral hindbrain sections showing 5-HT immunostaining (brown) after whole-mount X-gal staining of FEV2.2ΔZ (F) or FEV2.2Z (G) E12.5 embryos. Section shown in F is from the FEV2.2ΔZ founder embryo shown in E.
We next asked whether proximal conserved sequences were necessary for 5-HT neuron-specific expression by testing a transgene, FEV2.2ΔZ, in which sequences −214 to +25 of the proximal block were deleted in the context of the 2.2 kb upstream fragment (Fig. 1). Transgenic founders were analyzed at E12.5 using whole-mount X-gal staining. Deletion of these sequences led to a complete loss of serotonergic expression of FEV2.2ΔZ, although ectopic expression was present (Fig. 3E,F vs G). Similar results were obtained for a second transgene, FEV1.1ΔZ, in which −214/+25 sequences were removed from FEV1.1Z (Fig. 1).
Our findings show that proximal conserved sequences are not only sufficient but also necessary for 5-HT neuron expression of FEV in the developing rostral and anterior portion of the caudal hindbrain. Furthermore, they suggest that the FEV promoter comprises the 5′ flanking proximal sequences that are deleted in FEV2.2ΔZ and FEV1.1ΔZ. Consistent with this hypothesis, a 2.2 kb FEV transgene lacking the β-globin TATA box, FEV2.2ZΔβg, was expressed in a 5-HT neuron-specific pattern (Fig. 1). Therefore, the β-globin start site was not required for, and likely did not contribute to, transgene expression. Because FEV sequences can initiate transcription in the absence of a heterologous start site and deletion of proximal sequences results in loss of specific expression, we conclude that these conserved sequences immediately upstream of the FEV start site constitute the FEV promoter region.
Conserved GATA binding sites are present in the FEV promoter region
We next sought to identify putative transcription factor binding sites within the FEV upstream region to begin to determine factors that directly control FEV transcription. Upstream sequences were analyzed for candidate binding sites using the rVista (Loots and Ovcharenko, 2004) and MatInspector (Cartharius et al., 2005) tools. Interestingly, this analysis identified possible binding sites for NUDR/DEAF-1, Engrailed-1, and GATA transcription factors clustered in the promoter region (Fig. 4A). NUDR/DEAF-1 has been shown to be a repressor of the 5-HT1A receptor gene (Htr1a) through binding to an upstream polymorphic site (Lemonde et al., 2003), and loss-of-function studies have implicated Engrailed-1/2 in the development of 5-HT neurons (Simon et al., 2005). Of particular interest was the presence of two consensus GATA binding motifs (Fig. 4A,B). Multispecies comparisons of the upstream region revealed that these sites were conserved even among distantly related mammals (Fig. 4B). Because Gata-2 is essential for Pet-1 induction in the hindbrain (Craven et al., 2004), the presence of conserved tandem GATA sites in the FEV promoter region suggested that Gata-2 might directly induce Fev/Pet-1 in serotonergic precursors. Therefore, we focused our next set of studies at testing this idea.
Figure 4.
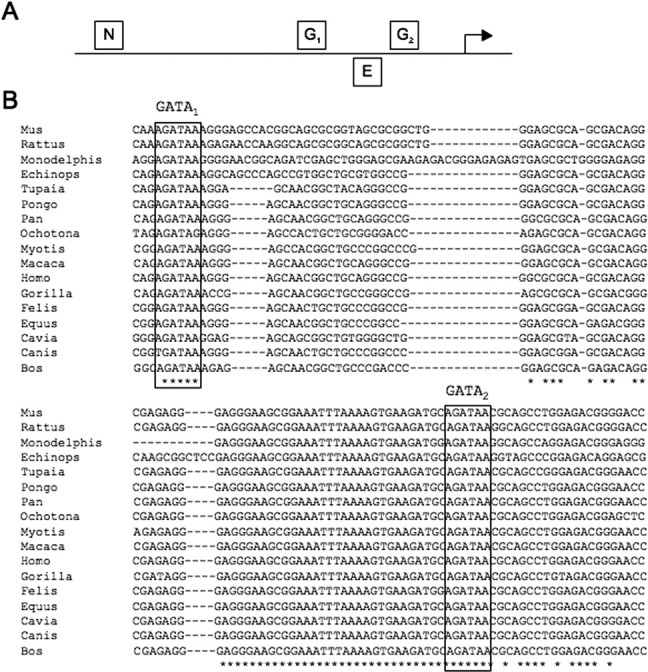
Predicted transcription factor binding sites in the FEV upstream region. A, Schematic showing approximate location of predicted binding sites for transcription factors implicated in serotonergic development. N, NUDR/DEAF1 at −202; G1 and G2, GATA sites GATA1 at −144 and GATA2 at −61, respectively; E, ENGRAILED-1 at −80. Coordinates are relative to the FEV transcriptional start site. B, Alignment of proximal sequences from 17 mammalian species. Predicted conserved GATA1 and GATA2 sites are boxed.
We first determined with coimmunostaining that Gata-2 was expressed in β-gal+ cells of the rostral hindbrain in FEV2.2Z transgenic embryos (Fig. 5A). We then asked whether the consensus sites could bind GATA protein in vitro. Gata-1, Gata-2, and Gata-3 all recognize a consensus site, WGATAR (Ko and Engel, 1993), and both sites in the FEV upstream region perfectly match this sequence (Fig. 4B). We tested the ability of purified recombinant Gata-1 protein to bind either the FEV distal (GATA1) or proximal (GATA2) sites (Fig. 4) using electrophoretic mobility shift assays (EMSAs). Recombinant Gata-1 protein formed a single complex with duplex oligonucleotides carrying either GATA site (Fig. 5B), thus demonstrating that these are indeed bona fide in vitro GATA binding sites. To test whether these sites are also recognized by Gata-2, we performed EMSAs with HeLa cell nuclear extracts, because these cells express endogenous Gata-2 but not other GATA factors (Dorfman et al., 1992; Zon et al., 1993; Siltanen et al., 1999; Ho et al., 2005). Oligonucleotides containing the distal conserved GATA site, GATA1, formed two complexes when incubated with HeLa nuclear extracts (arrow and arrowhead in Fig. 5C, lanes 2 and 5). These complexes were competed away with an excess of the unlabeled GATA1 site but not an equimolar mass of an oligonucleotide in which the GATA site had been altered (Fig. 5C, lanes 3 and 4, respectively). When the proximal GATA site, GATA2, was used as a competitor, wild-type but not mutant oligonucleotides specifically competed the upper complex formed with GATA1 oligonucleotides (Fig. 5C, lanes 6 and 7). GATA2 oligonucleotides formed similar complexes with HeLa extract that were competed with wild-type but not mutated GATA1 oligonucleotides (data not shown). These cross-competition experiments demonstrate that common proteins bind specifically to the GATA motif found in both oligonucleotide sequences. Finally, the addition of anti-Gata-2 antibody, but not an equal amount of anti-GFP antibody, disrupted the upper complex, indicating that Gata-2 is indeed present in that complex (Fig. 5D). Together, these results suggest that both sequences are functional in vitro binding sites for Gata-2.
Figure 5.
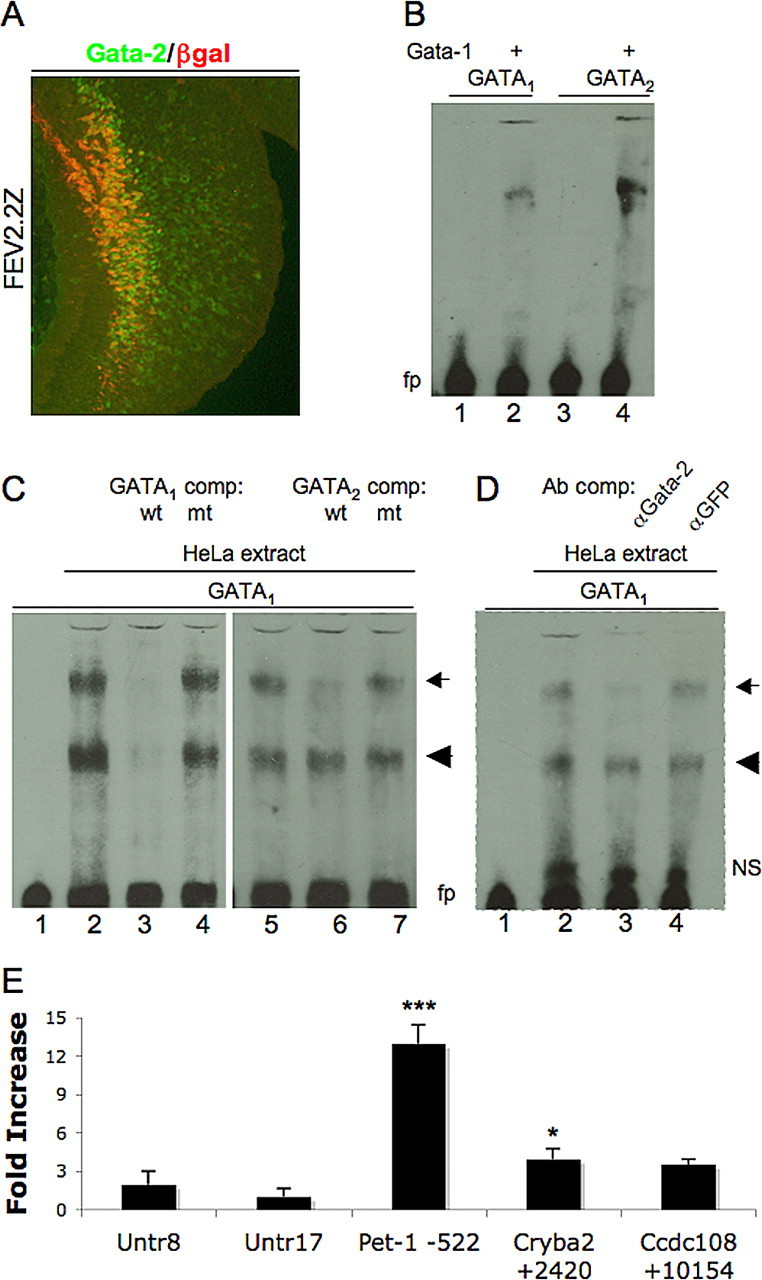
In vitro and in vivo analysis of GATA sites in FEV/Pet-1 upstream region. A, Coimmunostaining of sagittal section from rostral hindbrain of E11.5 FEV2.2Z embryo for Gata-2 and β-gal. B, Mobility shift assays with recombinant Gata-1 protein and 80 fmol of FEV biotinylated duplex oligonucleotides containing either GATA1 (lanes 1, 2) or GATA2 sequences (lanes 3, 4). C, Mobility shift assays with 0 (lane 1) or 12.8 μg of HeLa cell extract (lanes 2–7) and 80 fmol of GATA1 oligonucleotides. Lanes 3, 4, Preincubation with 100-fold excess of unlabeled GATA1 competitor (lane 3) or substituted GATA1 (lane 4) duplex oligonucleotides. Lanes 6, 7, Preincubation with 100-fold excess of unlabeled GATA2 competitor (lane 6) or substituted GATA2 (lane 7) duplex oligonucleotides. D, Antibody perturbation assays with 60 fmol of GATA1 oligonucleotides and 0 (lane 1) or 6.4 μg of HeLa cell nuclear extract (lanes 2–4). Lane 3, Preincubation with anti-Gata-2 antibody. Lane 4, Preincubation with anti-GFP antibody. fp, Free probe; NS, nonspecific complex; wt, wild-type GATA sequence; mt, mutant GATA sequence. Arrow, Specific GATA-factor-containing complex; arrowhead, non-GATA-factor-containing specific complex. E, Chromatin immunoprecipitation for Gata-2, showing occupancy of indicated genomic regions in embryonic mouse hindbrain. Values represent fold increase in binding plus SD to the indicated regions compared with negative control region (Untr17). Untr, Untranscribed genomic region; Ccdc108, coiled-coil domain-containing 108; Cryba2, βA2 crystallin. *p < 0.05 for Cryba2 +2420 versus Untr17; ***p < 0.001 for Pet-1 −522 versus any other tested region, one-way ANOVA with Bonferroni's multiple comparison test.
Gata-2 directly interacts with the Pet-1 cis-regulatory region
We next used chromatin immunoprecipitation (ChIP) to determine whether Gata-2 occupies the homologous Pet-1 cis-regulatory region in vivo. To our knowledge, there is no neuronal cell line that expresses high levels of Pet-1 or FEV together with Gata-2. Therefore, we developed a ChIP assay with E11.5 mouse hindbrain as a source of serotonergic chromatin. Chromatin was immunoprecipitated with an anti-Gata-2 antibody, and the immunoprecipitate was assessed by quantitative PCR. We tested for Gata-2 interaction with the Pet-1 upstream region, as well as sequences located in upstream genes, Cryba2 and Ccdc108, in which consensus GATA motifs are located. Anti-Gata-2 antibody immunoprecipitation of sequences associated with the Cryba2 GATA site was slightly increased over background levels measured with the Untr17 control but not with the Untr8 control. No enrichment of Ccdc108 sequences was detected when compared with either of these controls (Fig. 5E). In contrast, binding of Gata-2 to the Pet-1 upstream region was highly significantly enriched (p < 0.001) compared with both negative control regions and the Cryba2 or Ccdc108 regions (Fig. 5E). These findings show that, in the developing hindbrain, Gata-2 directly binds to the FEV/Pet-1 upstream cis-regulatory region.
Conserved GATA binding sites in the FEV cis-regulatory region are required for serotonergic transgene expression
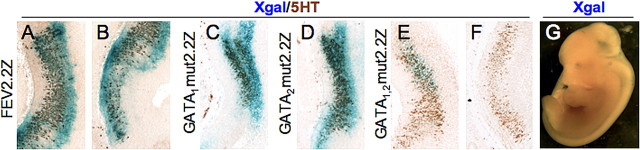
Having shown that conserved GATA binding sites are present in the FEV promoter region and that Gata-2 directly interacts with the homologous Pet-1 cis-regulatory region, we next investigated the function of the GATA sites in vivo. Site-directed mutagenesis was performed to destroy these GATA binding sites in FEV2.2Z, either singly (GATA1mut2.2Z or GATA2mut2.2Z) or in combination (GATA1,2mut2.2Z) (Fig. 1). Mutation of either GATA site alone resulted in little change in the transgene expression in 5-HT neurons (compare 6C, D with 6A) or reproducibility of expression; however, we did observe an increase in ectopic expression compared with FEV2.2Z or FEV2.2ZΔβg (data not shown). In contrast, nucleotide changes to both GATA sites (GATA1,2mut2.2Z) (Fig. 1) led to a complete loss of ventral hindbrain expression in 16 of 27 founders. The remaining 11 founders showed very weak expression in rostral 5-HT neurons (Fig. 6E,G), which was often accompanied by weak to strong ectopic expression. Little or no staining was observed in more median or caudal 5-HT neurons (Fig. 6, compare F, B). Furthermore, robust staining, comparable with FEV2.2Z (Figs. 2A, 6A), was never observed with GATA1,2mut2.2Z. Thus, mutation of both GATA sites greatly compromised the ability of the 2.2 kb fragment to direct expression to 5-HT neurons, as evidenced by both the large numbers of founders showing no hindbrain expression and the dramatically reduced serotonergic expression in remaining founders.
Figure 6.
GATA sites are functionally required for FEV transgene expression. A–F, Whole-mount X-gal staining of E12.5 transgenic embryos, followed by 5-HT immunostaining of sagittal slices from a FEV2.2Z embryo (A, B) compared with single GATA1 (C), GATA2 (D), or double GATA1,2 site mutations (E, F). A and C–E are of rostral 5-HT neurons. B and F show median 5-HT neurons. G, Whole-mount X-gal staining of E12.5 GATA1,2mut2.2Z founder. Sections of founder in G shown in E and F.
FEV-directed transgene expression and fate of mutant 5-HT neuron precursors in Pet-1−/− brain
We showed previously that a LacZ reporter controlled by the mouse Pet-1 upstream enhancer was weakly expressed in the Pet-1−/− brain compared with its expression in wild-type mice (Scott et al., 2005). To determine whether FEV-directed transgene expression was also dependent on endogenous Pet-1 function, we crossed the FEV2.2Z transgene into the Pet-1−/− background. The number of β-gal+ cells in the embryonic brain of FEV2.2Z Pet-1−/− mice was somewhat diminished compared with their numbers in Pet-1+/− background (Fig. 7A vs B), but FEV2.2Z continued to be expressed in 5-HT+ cells (Fig. 7A′, B′). However, FEV2.2Z expression was greatly diminished in adult Pet-1−/− mice (Fig. 7C vs D). When costained for TPH and β-gal, only a few colabeled cells were detected (Fig. 7E, arrowheads). These findings show that maintenance of FEV2.2Z expression depends on endogenous Pet-1.
Figure 7.
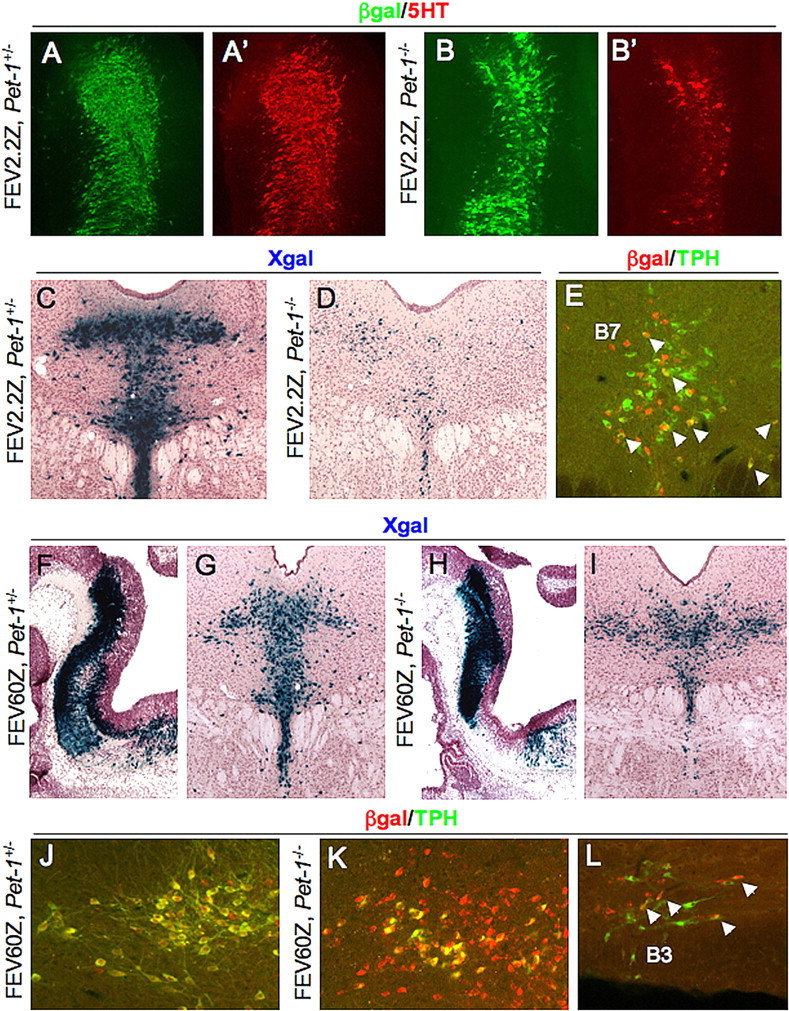
FEV-directed transgenes mark Pet-1−/− cells in the adult brain. A–B′, Coimmunostaining for β-gal and 5-HT in sagittal sections of the rostral hindbrain for FEV2.2Z, Pet-1+/− (A, A′) or FEV2.2Z, Pet-1−/− (B, B′) E12.5 embryos. C, D, X-gal staining of coronal sections from adult FEV2.2Z, Pet-1+/− (C) or FEV2.2Z, Pet-1−/− (D) mice. E, β-Gal/TPH coimmunostaining of a DRN section from a FEV2.2Z, Pet-1−/− adult mouse. F–I, X-gal staining of FEV60Z, Pet-1+/− (F, G) and FEV60Z, Pet-1−/− (H, I) at E12.5 (F, H) or adult (G, I) stages. J, K, Coimmunostaining for TPH and β-gal in FEV60Z, Pet-1+/− (J) or FEV60Z, Pet-1−/− (K, L) lateral B7 DRN (J, K) or B3 ventrolateral medulla (L). Arrowheads in E and L indicate colabeled cells.
To determine whether a longer upstream fragment was less sensitive to loss of Pet-1, we made a new reporter, FEV60Z, with a 60 kb upstream FEV fragment. Two independent FEV60Z transgenic lines were crossed into the Pet-1−/− background. In wild-type mice, we observed strong FEV60Z expression in the embryonic hindbrain at E12.5 (Fig. 7F and data not shown) and in adult DRN (Fig. 7G and data not shown). In contrast to FEV2.2Z and mouse Pet-1 enhancer-directed LacZ (Scott et al., 2005), the number of LacZ+ cells in the DRN was similar in FEV60Z, Pet-1−/− and FEV60Z, Pet-1+/− mice at both embryonic (Fig. 7F vs H) and adult (Fig. 7G vs I) stages. Indeed, cell counts of adult B7 DRN from Pet-1+/− and Pet-1−/− animals (n = 3 for each genotype, two sections per animal) revealed no differences between groups (means were 380 and 378 cells for Pet-1+/− and Pet-1−/− sections, respectively; t test p = 0.98).
In FEV60Z, Pet-1+/− mice, all TPH+ cells were β-gal+, and the majority of β-gal+ cells were TPH+ (Fig. 7J). However, in FEV60Z, Pet-1−/− mice, although all remaining TPH+ neurons in the B7 DRN expressed the transgene, a substantial number of β-gal+ cells were TPH− (Fig. 7K). In medullary raphe, FEV60Z was expressed in Pet-1−/− mice but not in all remaining TPH+ cells (Fig. 7L and data not shown). Thus, FEV60Z does not require Pet-1 to maintain its B7 expression but may be dependent on Pet-1 function in medullary 5-HT neurons.
The comparable expression of FEV60Z in Pet-1+/− and Pet-1−/− B7 DRN suggested that it could be used a novel fate marker of Pet-1−/− 5-HT neuron precursors. We reported previously (Hendricks et al., 2003) and confirmed here with FEV60Z (Fig. 7H,I,K) that normal numbers of B7 and B8 serotonergic precursors are generated in Pet-1−/− mice, but only ∼30% of them can be detected in the adult brain with traditional serotonergic markers. These observations suggested that cell death, misfating, or mismigration might account for the failure to detect these cells in adults. However, the similar numbers of β-gal+ cells in the adult B7 DRN of FEV60Z, Pet-1+/− and FEV60Z, Pet-1−/− mice indicated that Pet-1−/− mutant cells are not eliminated by a cell death program but evidently remain in an arrested precursor stage. The continued activity of FEV60Z is consistent with the maintenance of serotonergic character despite the lack of TPH expression in these cells (Fig. 7K). Additional studies showed that all FEV60Z+, Pet-1−/− DRN cells coexpressed NeuN (Fig. 8A), a general neuronal marker. The expression of Gata-3, which marks several distinct neuronal populations in the brain including postmitotic 5-HT neuron precursors (Pattyn et al., 2004; Zhao et al., 2008), was also present in β-gal+ cells (Fig. 8B,B′), further supporting a serotonergic character of FEV60Z+ cells in the Pet-1−/− brain. Importantly, these cells did not coexpress markers of nearby neuronal cell types, such as TH (Fig. 8C), a marker of dopaminergic neurons, or ChAT (Fig. 8D), expressed by motor neurons.
The marking of mutant 5-HT neuron precursors with the FEV60Z transgene also provided a means to visualize the organization of these cells in the adult B7 DRN. Interestingly, analysis of serial sections through the entire anteroposterior length of the B7 DRN suggested an aberrant distribution of β-gal+ cell bodies. The stereotypical “fountain” shape distribution of serotonergic cell bodies in the DRN was never observed in Pet-1-deficient mice, and β-gal+ cells in the nucleus appeared laterally extended (Fig. 7G vs I). Lateralization of the DRN in the Pet-1−/− brain was quantified by counting β-gal+ cells in two pairs of regions on each side of the midline (Fig. 8E, boxed areas). Significantly more β-gal+ cells were located farther from the midline in Pet-1−/− animals (Fig. 8F) than in controls. This lateralization appeared to be limited to DRN, because median and caudal β-gal+ cells were appropriately positioned. These findings suggest that Pet-1 may also be required for the precise migration of 5-HT neurons to their proper locations in the DRN.
Discussion
Our study of human FEV cis-regulatory control in mouse 5-HT neurons has led to three major findings. First, we show that a conserved region extending only 275 bp upstream of FEV is sufficient to direct transgene expression to 5-HT neuron precursors, whereas sequences farther upstream are likely required for maintenance of transcription in adult 5-HT neurons. Second, serotonergic expression of FEV/Pet-1 depends on direct interactions with the zinc finger factor Gata-2. Third, FEV transgenes mark serotonergic precursors in the adult brain and show that mutant cells remain in the Pet-1-deficient B7 DRN but their organization is disrupted. Thus, our findings provide new insights into the mechanisms that regulate FEV/Pet-1 in the ventral hindbrain and the roles of these factors in 5-HT neuron development.
Transcriptional control of FEV/Pet-1 in 5-HT neurons
Our data suggest that several regulatory subregions are responsible for different spatial and temporal domains of FEV transcription in 5-HT neurons. A proximal subregion surrounding the FEV transcriptional start site was sufficient for 5-HT neuron-specific expression of FEV transgenes in the rostral and the anterior half of the caudal hindbrain. The FEV promoter likely comprises sequences in the proximal subregion, because deletion of this region completely abolished transgene expression in developing 5-HT neurons. The proximal subregion, however, was not able to direct transgene expression to the posterior half of the caudal hindbrain, nor was it able to efficiently direct transgene expression to adult 5-HT neurons. Additional findings suggest that a separate regulatory module directs FEV transcription to the posterior hindbrain and another one functions to maintain FEV transcription. First, FEV2.2Z and smaller transgenes were unable to fully recapitulate the expression patterns of endogenous Pet-1 in the caudal hindbrain. However, FEV60Z was expressed in 5-HT neurons along the entire length of the caudal domain (data not shown), which suggests that a spatial regulatory module resides farther upstream of the immediate 2.2 kb 5′ flanking region. Second, FEV2.2Z but not FEV0.6Z was strongly expressed in adult 5-HT neurons. Therefore, the temporal module that functions to maintain FEV transcription in the adult dorsal and median raphe is likely to be located in or near the distal block of conserved sequences. These findings suggest that FEV/Pet-1 expression along the rostrocaudal axis of the developing hindbrain and in the adult brain may be controlled by heterogeneous cis-regulatory elements, each responsible for different spatial and temporal domains of FEV transcription.
Direct Gata-2 regulation of FEV/Pet-1
Before this study, it was not clear which, if any, of the transcription factors implicated in the serotonergic regulatory cascade are direct activators of Pet-1 or FEV. Previous experiments placed Gata-2 genetically upstream of Pet-1 at all rostrocaudal levels of the developing hindbrain: Pet-1 expression and all 5-HT neurons are lost in Gata-2 knock-outs, whereas misexpression of Gata-2 in chick r1 induces Pet-1 and ectopic 5-HT neurons (Craven et al., 2004). Our own studies expand on these findings to support direct Gata-2–FEV/Pet-1 interactions. First, we identify two conserved GATA binding sites (Figs. 4, 5) in the FEV promoter region that are recognized in vitro by GATA factors and, in particular, Gata-2. Second, a ChIP assay performed on mouse embryonic hindbrain shows that Gata-2 protein directly interacts with the Pet-1 cis-regulatory region. Third, an FEV transgene lacking both GATA sites (GATA1,2mut2.2Z) showed weak or undetectable expression in developing 5-HT neurons but increased transgene expression in nonserotonergic cell types. Elimination of each GATA site individually had little effect on transgene expression in 5-HT neurons, which suggests the GATA sites are functionally redundant for enhancement of FEV transcription in 5-HT neurons. Interestingly, the related factor Gata-3 is not redundant with Gata-2, because the loss of Pet-1 expression in Gata-2 knock-outs occurs despite the continued expression of Gata-3 (Craven et al., 2004). Additionally, Pet-1 expression is unchanged in Gata-3 knock-out mice, in which a subset of 5-HT neurons is lost (Pattyn et al., 2004), suggesting that, although Gata-3 plays a role in 5-HT neuron development, it is unlikely to do so via regulation of Pet-1.
The weak serotonergic expression of GATA1,2mut2.2Z seen in some lines contrasts with the complete absence of transgene expression observed in all lines after deletion of the proximal sequences that included both GATA sites. Therefore, additional transcription factor interactions are also likely to occur in the conserved promoter region and are probably involved in restricting expression of FEV to 5-HT neurons because GATA-2 is expressed in many types of neurons. We suspect that Gata-2 interactions with sites in the FEV promoter region are not likely to be involved in maintenance of expression in the adult brain because Gata-2 expression declines around the time of Pet-1 induction (Nardelli et al., 1999; Zhou et al., 2000), a finding consistent with the lack of FEV0.6Z reporter expression in adult 5-HT neurons. Our findings provide insight into the 5-HT neuron developmental program by showing that FEV and Pet-1 are direct targets of a conserved Gata-2-dependent regulatory cascade that gives rise to 5-HT neurons in the developing rostral and caudal hindbrain.
Fate of mutant 5-HT neuron precursors in the Pet-1−/− brain
Our FEV cis-regulatory studies have identified a unique marker of Pet-1−/− cells in the B7 DRN, which has led to new insights into the role of Pet-1 in 5-HT neuron development. In adult Pet-1−/− mice, some cell bodies are missing in the mutant pons in which 5-HT neurons normally coalesce to form the B6 DRN (Hendricks et al., 2003). Therefore, cell death or misfating may account for the absence of some 5-HT neurons in the Pet-1−/− pons. However, the large numbers of FEV60Z+ cells in the Pet-1−/− B7 DRN and B8 MRN suggests that the majority of mutant precursors neither die nor misfate but rather are maintained in an arrested state of development in which coordinate transcription of TPH2, aromatic amino acid decarboxylase, SERT, and vesicular monoamine transporter 2 is greatly reduced or aborted. In support of this idea, NeuN immunostaining showed that FEV60Z+ mutant precursors retained pan-neuronal character. In addition, they maintained expression of Gata-3, a postmitotic (although not specific) marker of serotonergic precursors. In contrast, nearly all 5-HT neuron cell bodies were shown to be absent in adult mice in which Lmx1b was conditionally inactivated in the serotonergic lineage (Zhao et al., 2006). Thus, whereas Pet-1 and Lmx1b are likely to cooperate in the coordinate transcriptional control of serotonergic gene expression and 5-HT synthesis (Cheng et al., 2003), Lmx1b appears to function independently from Pet-1 in the survival of 5-HT neurons. It is unknown whether mutant 5-HT neuron precursors in the adult Pet-1−/− B7 DRN retain physiologic functions such as release of neuropeptides. Additional studies facilitated by the genetic marking of these cells with FEV60Z will likely resolve this issue.
An unexpected finding of the FEV60Z studies was the altered distribution of cells in the Pet-1−/− DRN. These findings suggest that Pet-1 function may be required for precise migration patterns of 5-HT neurons. The migration of 5-HT precursors is also deficient in Lmx1b−/− mice (Ding et al., 2003), suggesting that Pet-1 and Lmx1b may coordinately regulate migration, perhaps through regulation of 5-HT synthesis. 5-HT can modulate the in vitro migration of neural crest cells (Moiseiwitsch and Lauder, 1995) and is attractive for mast cells in vitro and in vivo (Kushnir-Sukhov et al., 2006). In both cell types, signaling through the Htr1a receptor was implicated. Htr1a expression is already enriched in newly differentiated 5-HT neurons as they begin to migrate (C. J. Wylie and E. S. Deneris, unpublished observations). This raises the possibility that release of 5-HT from embryonic 5-HT neurons provides an autoregulatory signal that helps direct migration of 5-HT precursors to their mature positions. It remains to be determined what might be the functional consequence of serotonergic cell body disorganization in the DRN and whether this defect contributes to the behavioral and physiological alterations present in Pet-1-deficient mice.
Transcriptional control of FEV and neuropsychiatric disease
Defects in the transcriptional control of 5-HT neuron gene expression lead to effects on adult emotional behavior (Hendricks et al., 2003), neonatal respiration (Erickson et al., 2007), thermoregulation (Hodges et al., 2008), inflammatory pain (Zhao et al., 2007), and reproductive success (Lerch-Haner et al., 2008). Furthermore, genetic variants that alter TPH2 and SERT expression have been associated with psychiatric disease (Leonardo and Hen, 2006; Murphy and Lesch, 2008). These findings raise the possibility that genetic alterations impacting serotonergic transcriptional control may predispose individuals to abnormal behavior or physiology (Albert and Lemonde, 2004). The ability of FEV to regulate TPH2 and SERT (Lerch-Haner et al., 2008) suggests the regulatory mechanisms governing FEV expression may be relevant to disease pathogenesis. Our studies provide a functional map of FEV cis-regulatory elements, which will facilitate the identification and analysis of possible FEV cis-regulatory variants and their roles as potential disease susceptibility factors. Indeed, the NCBI dbSNP database (Sherry et al., 2001) lists a possible regulatory variant (rs57981340) within the FEV promoter region we have defined here, and our sequencing has verified the existence of this SNP (K. C. Krueger, R. L. Findling, and E. S. Deneris, unpublished data).
Footnotes
This work was supported by National Institutes of Health Grant MH062723. We thank the Case Transgenic and Targeting Facility for generation of transgenic lines. We also thank Kathy Lobur and RoxAnne Murphy for outstanding assistance with propagation, maintenance, and genotyping of Pet-1−/− lines.
References
- Albert PR, Lemonde S. 5-HT1A receptors, gene repression, and depression: guilt by association. Neuroscientist. 2004;10:575–593. doi: 10.1177/1073858404267382. [DOI] [PubMed] [Google Scholar]
- Alexiadis V, Ballestas ME, Sanchez C, Winokur S, Vedanarayanan V, Warren M, Ehrlich M. RNAPol-ChIP analysis of transcription from FSHD-linked tandem repeats and satellite DNA. Biochim Biophys Acta. 2007;1769:29–40. doi: 10.1016/j.bbaexp.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge MS, Hen R, Gingrich JA. Neurodevelopmental origins of depressive disorders. Curr Opin Pharmacol. 2007;7:8–17. doi: 10.1016/j.coph.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, Zhang X, Rodriguiz RM, Sotnikova TD, Cools MJ, Wetsel WC, Gainetdinov RR, Caron MG. Role of GSK3 beta in behavioral abnormalities induced by serotonin deficiency. Proc Natl Acad Sci USA. 2008;105:1333–1338. doi: 10.1073/pnas.0711496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Cheng L, Chen CL, Luo P, Tan M, Qiu M, Johnson R, Ma Q. Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J Neurosci. 2003;23:9961–9967. doi: 10.1523/JNEUROSCI.23-31-09961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven SE, Lim KC, Ye W, Engel JD, de Sauvage F, Rosenthal A. Gata2 specifies serotonergic neurons downstream of sonic hedgehog. Development. 2004;131:1165–1173. doi: 10.1242/dev.01024. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- Dorfman DM, Wilson DB, Bruns GA, Orkin SH. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- Erickson JT, Shafer G, Rossetti MD, Wilson CG, Deneris ES. Arrest of 5-HT neuron differentiation delays respiratory maturation and impairs neonatal homeostatic responses to environmental challenges. Respir Physiol Neurobiol. 2007;159:85–101. doi: 10.1016/j.resp.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frengen E, Weichenhan D, Zhao B, Osoegawa K, van Geel M, de Jong PJ. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics. 1999;58:250–253. doi: 10.1006/geno.1998.5693. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES. The ETS domain factor Pet-1 is an early and precise marker of central 5-HT neurons and interacts with a conserved element in serotonergic genes. J Neurosci. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Ho CK, Wood JR, Stewart DR, Ewens K, Ankener W, Wickenheisser J, Nelson-Degrave V, Zhang Z, Legro RS, Dunaif A, McAllister JM, Spielman R, Strauss JF., 3rd Increased transcription and increased messenger ribonucleic acid (mRNA) stability contribute to increased GATA6 mRNA abundance in polycystic ovary syndrome theca cells. J Clin Endocrinol Metab. 2005;90:6596–6602. doi: 10.1210/jc.2005-0890. [DOI] [PubMed] [Google Scholar]
- Hodges MR, Tattersall GJ, Harris MB, McEvoy SD, Richerson DN, Deneris ES, Johnson RL, Chen ZF, Richerson GB. Defects in breathing and thermoregulation in mice with near-complete absence of central serotonin neurons. J Neurosci. 2008;28:2495–2505. doi: 10.1523/JNEUROSCI.4729-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyo AH, Porter B, Deneris ES, Austin MC. Regional distribution and cellular localization of the ETS-domain transcription factor, FEV, mRNA in the human postmortem brain. Synapse. 2005;57:223–228. doi: 10.1002/syn.20178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob J, Ferri AL, Milton C, Prin F, Pla P, Lin W, Gavalas A, Ang SL, Briscoe J. Transcriptional repression coordinates the temporal switch from motor to serotonergic neurogenesis. Nat Neurosci. 2007;10:1433–1439. doi: 10.1038/nn1985. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Engel JD. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi-Osaki M, Ohneda O, Suzuki N, Minegishi N, Yokomizo T, Takahashi S, Lim KC, Engel JD, Yamamoto M. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir-Sukhov NM, Gilfillan AM, Coleman JW, Brown JM, Bruening S, Toth M, Metcalfe DD. 5-hydroxytryptamine induces mast cell adhesion and migration. J Immunol. 2006;177:6422–6432. doi: 10.4049/jimmunol.177.9.6422. [DOI] [PubMed] [Google Scholar]
- Lemonde S, Turecki G, Bakish D, Du L, Hrdina PD, Bown CD, Sequeira A, Kushwaha N, Morris SJ, Basak A, Ou XM, Albert PR. Impaired repression at a 5-hydroxytryptamine 1A receptor gene polymorphism associated with major depression and suicide. J Neurosci. 2003;23:8788–8799. doi: 10.1523/JNEUROSCI.23-25-08788.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo ED, Hen R. Genetics of affective and anxiety disorders. Annu Rev Psychol. 2006;57:117–137. doi: 10.1146/annurev.psych.57.102904.190118. [DOI] [PubMed] [Google Scholar]
- Lerch-Haner JK, Frierson D, Crawford LK, Beck SG, Deneris ES. Serotonergic transcriptional programming determines maternal behavior and offspring survival. Nat Neurosci. 2008;11:1001–1003. doi: 10.1038/nn.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidov HG, Molliver ME. An immunohistochemical study of serotonin neuron development in the rat: ascending pathways and terminal fields. Brain Res Bull. 1982;8:389–430. doi: 10.1016/0361-9230(82)90077-6. [DOI] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucleic Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseiwitsch JR, Lauder JM. Serotonin regulates mouse cranial neural crest migration. Proc Natl Acad Sci USA. 1995;92:7182–7186. doi: 10.1073/pnas.92.16.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DL, Lesch KP. Targeting the murine serotonin transporter: insights into human neurobiology. Nat Rev Neurosci. 2008;9:85–96. doi: 10.1038/nrn2284. [DOI] [PubMed] [Google Scholar]
- Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210:305–321. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes Dev. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Simplicio N, van Doorninck JH, Goridis C, Guillemot F, Brunet JF. Ascl1/Mash1 is required for the development of central serotonergic neurons. Nat Neurosci. 2004;7:589–595. doi: 10.1038/nn1247. [DOI] [PubMed] [Google Scholar]
- Pfaar H, von Holst A, Vogt Weisenhorn DM, Brodski C, Guimera J, Wurst W. mPet-1, a mouse ETS-domain transcription factor, is expressed in central serotonergic neurons. Dev Genes Evol. 2002;212:43–46. doi: 10.1007/s00427-001-0208-x. [DOI] [PubMed] [Google Scholar]
- Rand CM, Berry-Kravis EM, Zhou L, Fan W, Weese-Mayer DE. Sudden infant death syndrome: rare mutation in the serotonin system FEV gene. Pediatr Res. 2007;62:180–182. doi: 10.1203/PDR.0b013e3180a725a0. [DOI] [PubMed] [Google Scholar]
- Scott MM, Krueger KC, Deneris ES. A differentially autoregulated Pet-1 enhancer region is a critical target of the transcriptional cascade that governs serotonin neuron development. J Neurosci. 2005;25:2628–2636. doi: 10.1523/JNEUROSCI.4979-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siltanen S, Anttonen M, Heikkilä P, Narita N, Laitinen M, Ritvos O, Wilson DB, Heikinheimo M. Transcription factor GATA-4 is expressed in pediatric yolk sac tumors. Am J Pathol. 1999;155:1823–1829. doi: 10.1016/S0002-9440(10)65500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon HH, Scholz C, O'Leary DD. Engrailed genes control developmental fate of serotonergic and noradrenergic neurons in mid- and hindbrain in a gene dose-dependent manner. Mol Cell Neurosci. 2005;28:96–105. doi: 10.1016/j.mcn.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Steinbusch HWM. Distribution of serotonin-immunoreactivity in the central nervous system of the rat-cell bodies and terminals. Neuroscience. 1981;6:557–618. doi: 10.1016/0306-4522(81)90146-9. [DOI] [PubMed] [Google Scholar]
- Wallace JA, Lauder JM. Development of the serotonergic system in the rat embryo: an immunocytochemical study. Brain Res Bull. 1983;10:459–479. doi: 10.1016/0361-9230(83)90144-2. [DOI] [PubMed] [Google Scholar]
- Yee SP, Rigby PW. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes Dev. 1993;7:1277–1289. doi: 10.1101/gad.7.7a.1277. [DOI] [PubMed] [Google Scholar]
- Zhao GY, Li ZY, Zou HL, Hu ZL, Song NN, Zheng MH, Su CJ, Ding YQ. Expression of the transcription factor GATA3 in the postnatal mouse central nervous system. Neurosci Res. 2008;61:420–428. doi: 10.1016/j.neures.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Zhao ZQ, Scott M, Chiechio S, Wang JS, Renner KJ, Gereau RW, 4th, Johnson RL, Deneris ES, Chen ZF. Lmx1b is required for maintenance of central serotonergic neurons and mice lacking central serotonergic system exhibit normal locomotor activity. J Neurosci. 2006;26:12781–12788. doi: 10.1523/JNEUROSCI.4143-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ZQ, Chiechio S, Sun YG, Zhang KH, Zhao CS, Scott M, Johnson RL, Deneris ES, Renner KJ, Gereau RW, 4th, Chen ZF. Mice lacking central serotonergic neurons show enhanced inflammatory pain and an impaired analgesic response to antidepressant drugs. J Neurosci. 2007;27:6045–6053. doi: 10.1523/JNEUROSCI.1623-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yamamoto M, Engel JD. GATA2 is required for the generation of V2 interneurons. Development. 2000;127:3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
- Zon LI, Yamaguchi Y, Yee K, Albee EA, Kimura A, Bennett JC, Orkin SH, Ackerman SJ. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: potential role in gene transcription. Blood. 1993;81:3234–3241. [PubMed] [Google Scholar]