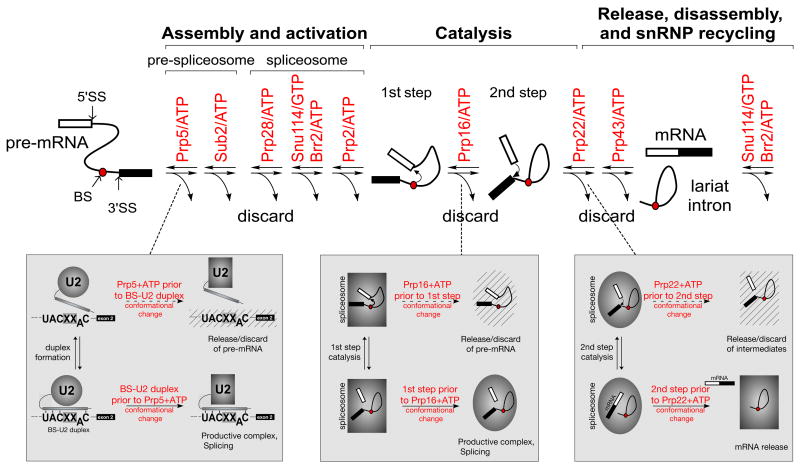
Figure 3. NTPase-associated steps during splicing offer opportunities for kinetic discrimination of suboptimal pre-mRNA substrates.
(upper) Schematic of transitions facilitated by DExD/H-box ATPases and the Snu114 GTPase during pre-mRNA splicing. SS, splice site; BS, branch site. (lower) Characterised examples of kinetic proofreading mediated by spliceosomal ATPases: (left) Altered competition between BS-U2 pairing and the conformational change mediated by the Prp5 ATPase changes the fidelity of BS selection. (center) Altered competition between the first catalytic step and Prp16 ATPase activity affects the fidelity of splice site usage in this step. (right) Altered competition between the second catalytic step and Prp22 ATPase affects second step splice site fidelity.