Fig. 1.
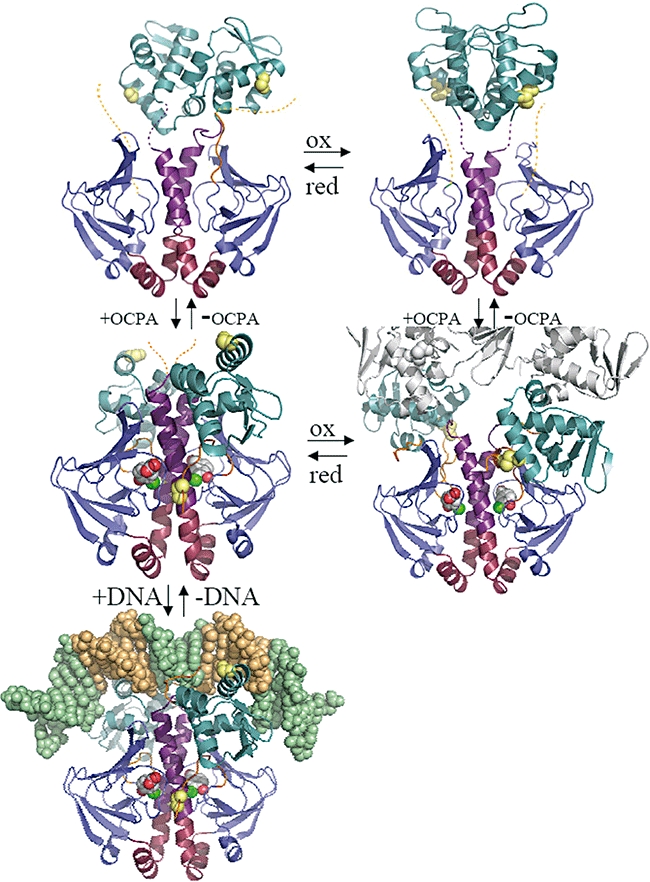
Atomic structures of D. hafniense CprK in different states. A representative dimer is depicted for each structure in cartoon form. Individual structural elements are coloured as follows, the N-terminus (residues 1–18) in orange, the effector β-barrel domain (19–107) in blue, the central α-helices not affected in position by ligand binding (108–127) in red, the central α-helix region affected by ligand binding + loop region in purple (128–148), the DNA-binding domain (149–227) in teal, the C-terminus (228–232) in orange. In addition, the position of C11 and C200 is indicated (when visible) by yellow spheres. The bound OCPA ligand is depicted in atom coloured spheres with grey carbons. The bound DNA is depicted in atom spheres with nucleotides constituting the (de)halobox sequence in pale orange. In case of the oxidized CprK:OCPA complex a symmetry-related molecule is depicted in grey to illustrate the changed quaternary structure in this particular state. Dotted lines indicate the position of highly mobile linker elements or N/C termini not visible in the electron density maps. Structures are organized as follows, top left: CprKC200S; top right: oxidized CprK; middle left: CprKC200S:OCPA; middle right: oxidized CprK:OCPA (previously determined 2H6B; Joyce et al., 2006); bottom left: (de)halobox DNA: CprKC200S:OCPA complex.
