Fig. 6.
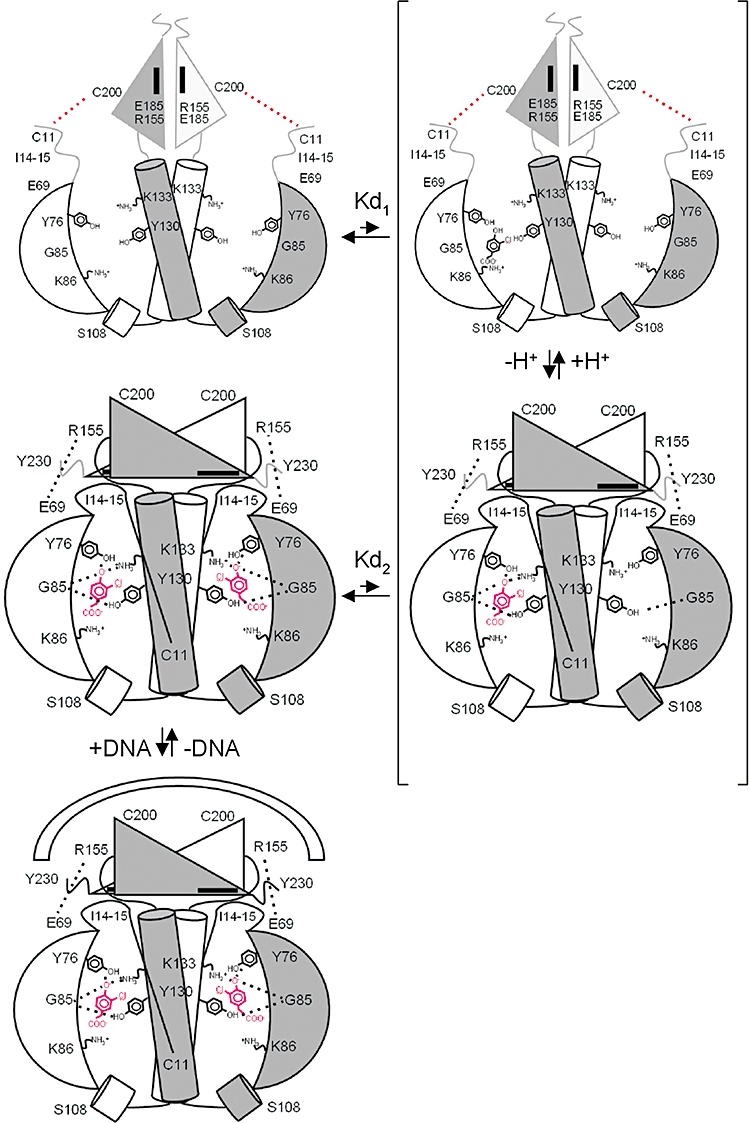
Model for allosteric effects of OCPA binding by CprK. A schematic model illustrating the extreme positive cooperativity model for OCPA binding by CprK and associated structural reorganization. Individual structural elements are depicted in grey scale when observed to be highly mobile. Key amino acids and contacts are indicated where appropriate. For clarity, the DNA-binding domain hydrophobic set of residues involved in domain interactions (Leu-156, Leu-160, Leu-178, Met-176, Ile-186) is depicted as a black rectangle. The bound OCPA molecules are depicted in black (protonated) or red (deprotonated). We postulate binding of the first ligand is characterized by weak binding (Kd1∼2.5 mM) but following ligand deprotonation (as predicted by our pKa interrogation model; Joyce et al., 2006) and concomitant reorganization of the entire CprK a high affinity ligand binding site is created for the second ligand (Kd2∼1 μm). It is unclear whether deprotonation occurs prior to or during binding for the second ligand molecule (here depicted as binding in the deprotonated form).
