Fig. 7.
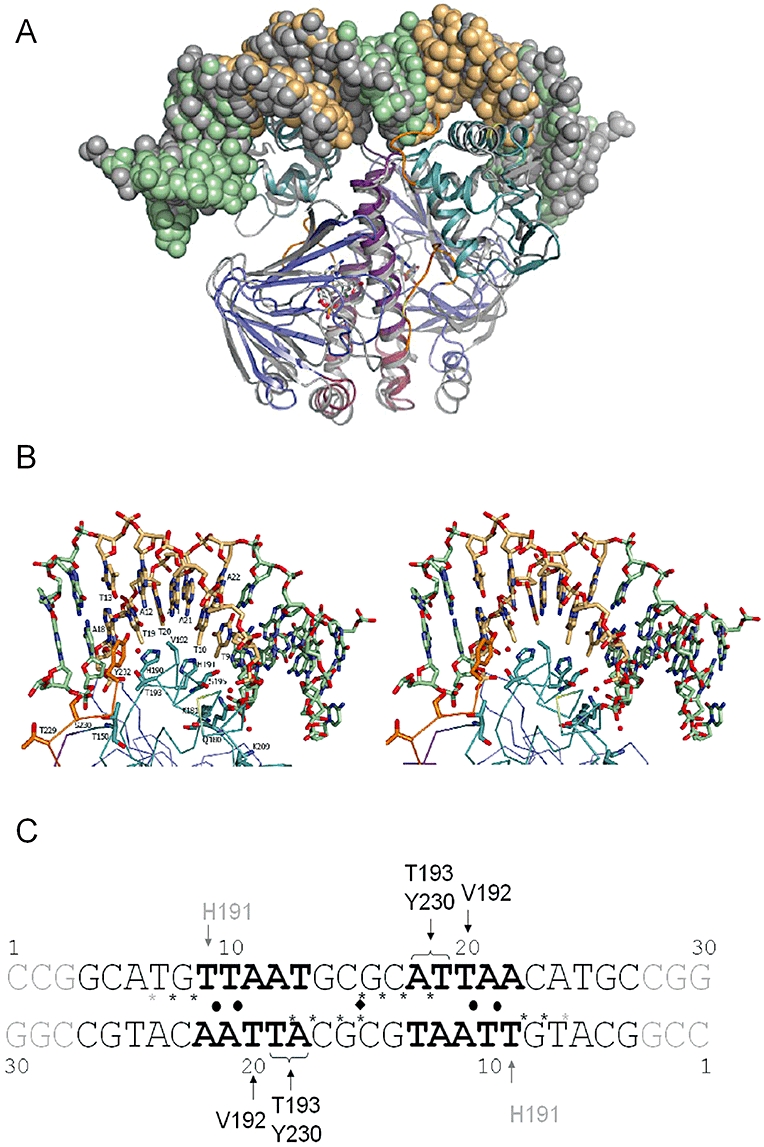
(de)halobox DNA binding by CprKC200S:OCPA.
A. An overlay of the CprKC200S:OCPA:DNA crystal structure with the available CRP:cAMP:DNA structure (PDB code 2CGP). The overlay is based on structural alignment of the respective HTH motives. CprK and associated DNA are depicted and colour coded as in Fig. 1. The CRP is similarly represented but in grey tones.
B. Stereoview of DNA–CprK interface contact area. The DNA and key amino acids are represented in atom coloured sticks with carbons coloured according to Fig. 1.
C. Schematic overview of the various CprKC200S DNA contacts established. The (de)halobox consensus sequence is depicted in bold while nucleotides that were not observed in the structure are depicted in grey scale. A black sphere indicates the base pairs for which geometric parameters deviate significantly from average. DNA backbone phosphates that make direct contact with CprK residues are depicted by a star.
