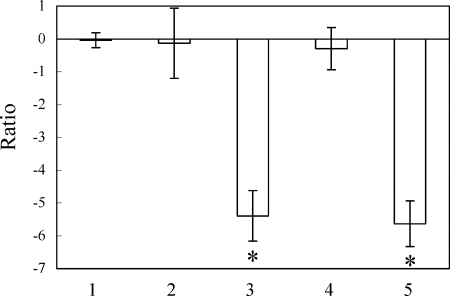
FIG. 2.
EMA and real-time PCR combined analyses of viable or dead cells of L. pneumophila strain 80-045. The number of bacteria was estimated from the amount of DNA detected by real-time PCR with SYBR green as the reporter dye. The number of viable cells without EMA treatment was set as 1. The numbers of EMA-treated viable cells (bar 1), untreated heat-killed cells (bar 2), EMA-treated heat-killed cells (bar 3), untreated chlorine-killed cells (bar 4), and EMA-treated chlorine-killed cells (bar 5) are described as ratios against the number of untreated viable cells [i.e., ratio = log10 (number of test cells/number of untreated viable cells)]. The error bars represent standard deviations from more than three independent experiments. Asterisks indicate significant decreases in the numbers of EMA-treated samples compared to those of untreated samples.