Abstract
To examine the role of the PmrA/PmrB two-component system (TCS) of Legionella pneumophila in global gene regulation and in intracellular infection, we constructed pmrA and pmrB isogenic mutants by allelic exchange. Genome-wide microarray gene expression analyses of the pmrA and pmrB mutants at both the exponential and the postexponential phases have shown that the PmrA/PmrB TCS has a global effect on the expression of 279 genes classified into nine groups of genes encoding eukaryotic-like proteins, Dot/Icm apparatus and secreted effectors, type II-secreted proteins, regulators of the postexponential phase, stress response genes, flagellar biosynthesis genes, metabolic genes, and genes of unknown function. Forty-one genes were differentially regulated in the pmrA or pmrB mutant, suggesting a possible cross talk with other TCSs. The pmrB mutant is more sensitive to low pH than the pmrA mutant and the wild-type strain, suggesting that acidity may trigger this TCS. The pmrB mutant exhibits a significant defect in intracellular proliferation within human macrophages, Acanthamoeba polyphaga, and the ciliate Tetrahymena pyriformis. In contrast, the pmrA mutant is defective only in the ciliate. Despite the intracellular growth defect within human macrophages, phagosomes harboring the pmrB mutant exclude late endosomal and lysosomal markers and are remodeled by the rough endoplasmic reticulum. Similar to the dot/icm mutants, the intracellular growth defect of the pmrB mutant is totally rescued in cis within communal phagosomes harboring the wild-type strain. We conclude that the PmrA/PmrB TCS has a global effect on gene expression and is required for the intracellular proliferation of L. pneumophila within human macrophages and protozoa. Differences in gene regulation and intracellular growth phenotypes between the pmrA and pmrB mutant suggests a cross talk with other TCSs.
Legionella pneumophila, the causative agent of Legionnaires' disease, is an intracellular bacterium that replicates within protozoa and human macrophages (40, 43, 75). Protozoa are the primary hosts of L. pneumophila in the natural aquatic environment (3, 7, 36, 40, 51, 58, 73). Infection of the human host is considered an accidental diversion from the natural life cycle within protozoa (36, 51). When water aerosol containing L. pneumophila is inhaled or contaminated water is aspirated, L. pneumophila enters the human lung and infects alveolar macrophages and epithelial cells, leading to an atypical pneumonia known as Legionnaires' disease (76, 77). After entry, the Legionella-containing phagosomes evade the default endocytic traffic and intercept endoplasmic reticulum (ER) vesicles to establish a replicative niche (16, 42, 45, 59, 70, 74).
Governed by a biphasic life cycle within amoeba host, L. pneumophila alternates between a replicative form and a mature intracellular form that is highly infectious to cells and resistant to environmental stress (28, 29, 33, 38). In vitro, this phenotypic modulation triggered upon transition from the exponential (E) to the postexponential (PE) phase requires a delicate regulatory cascade that can be triggered by nutrient limitation (28, 33, 38). At the PE phase, L. pneumophila relies on two ppGpp synthases, RelA and SpoT, both of which are essential for differentiation and phenotypic modification at the PE phase. Synthesis of ppGpp in response to amino acid starvation is RelA dependent (33, 34). Whereas relA mutant had no defective phenotype in macrophages, the relA/spoT double mutant is totally defective. The accumulation of the alarmone molecule ppGpp stimulates the LetA/LetS two-component system (TCS), the sigma factors RpoS, RpoN, RpoD, and FliA, and the mRNA-binding repressor protein (CsrA), leading to a phenotypic switch from the intracellular replicative form to the transmissive form (28, 33, 34, 38, 52, 61, 80).
The Dot/Icm type IV secretion system, which is encoded by 26 genes, is required for phagosome biogenesis and intracellular proliferation (27, 63, 64). L. pneumophila modulates the trafficking of its phagosome via the action of Dot/Icm-translocated effector proteins (19, 45, 46, 71). The regulation of expression of genes encoding both the Dot/Icm apparatus and some of its substrates has been proposed to be mediated in part by the regulatory cascades triggered at the PE phase (25). Recent work has shown a role for the PmrA/PmrB TCS in the regulation of expression of several genes encoding Dot/Icm-secreted effectors in L. pneumophila (79). The PmrA/PmrB TCS is a bacterial signal transduction system that mediates bacterial responses to various stimuli (39), which may be biotic or abiotic and may be triggered via quorum sensing (37). This TCS consists of a membrane-bound sensor protein (PmrB) that monitors the environment and responds to a specific signal (23) to activate a cognate response regulator protein (PmrA). The response regulator then recognizes and binds to a specific DNA sequence, leading to the modulation of transcription (23). The number of TCSs in L. pneumophila is substantially lower than in other bacteria such as Escherichia coli, which was estimated to harbor 40 different sensor-regulator pairs (49). The PmrA/PmrB TCS is conserved in all four published genomes of L. pneumophila: Lens, Paris, Corby, and Philadelphia-1 (12-15, 65). In Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa, the PmrA/PmrB system has been shown to regulate genes that modify lipopolysaccharide and confer bacterial resistance to cationic antimicrobial peptides and polymyxin B and is triggered in response to limiting Mg2+ conditions, high levels of Fe3+, and low pH (31, 57). PmrA was identified as a major regulator of the feoAB operon in P. aeruginosa and S. enterica serovar Typhimurium; this locus is known to be involved in iron acquisition and assimilation in L. pneumophila as well (22, 48, 56). In L. pneumophila, PmrA plays a role in regulating several Dot/Icm-secreted effectors (6, 21, 79), but the environmental signal activating PmrB is unknown.
Zusman et al. previously showed that the PmrA response regulator of L. pneumophila promotes the intracellular infection of HL-60 macrophages (79). However, the role of L. pneumophila PmrB in the intracellular infection, as well as in the regulation of expression of L. pneumophila virulence traits, remains unknown. We characterized here both the pmrA and the pmrB mutants of L. pneumophila. We show that PmrB is involved in the intracellular infection of macrophages and amoebas and that both PmrA and PmrB are necessary for the infection of ciliates. Despite its growth defect, the pmrB mutant is not required for evasion of the endocytic pathway, and its defect is totally rescued in the communal phagosome established by the wild-type (WT) strain. The pmrB mutant is more sensitive to acidic environments compared to the WT strain, suggesting that low pH may trigger the PmrA/PmrB TCS. Genome-wide microarray analyses suggest a central role for PmrA in the regulation of the L. pneumophila life cycle, and a possible cross talk between the PmrA/PmrB TCS and other L. pneumophila TCSs is proposed.
MATERIALS AND METHODS
DNA manipulations.
DNA manipulations and restriction enzyme digestions were performed by using standard procedures. Restriction enzymes and T4 DNA ligase were purchased from Promega (Madison, WI). L. pneumophila chromosomal DNA was prepared by using the Puregene DNA isolation kit from Gentra Systems (Minneapolis, MN). Plasmid preparations were performed with the Bio-Rad Quantum miniprep kit. Purification of DNA fragments from agarose gels for subcloning was carried out with a QIAquick gel purification kit (Qiagen, Inc., Valencia, CA). Fragments containing L. pneumophila pmrA and pmrB genes were cloned into the plasmid vector pBC-SK+ (Stratagene, Inc., La Jolla, CA), and the resulting clone was mutagenized by using an EZ-Tn5<KAN-2> in vitro transposome insertion kit from Epicentre. Transformation of E. coli strain DH5α by electroporation was performed with a BTX ECM 630, as recommended by Invitrogen Corp. (Carlsbad, CA). Mutations of the parental strain AA100 were carried out by allelic exchange with the kanamycin (Kan) insertion mutagenized pmrA and pmrB clone after natural transformation, as previously described (67, 68). The isogenic pmrA and pmrB mutants were transcomplemented with the same plasmid vector harboring the corresponding gene that was used as a template for mutagenesis. The primer pairs used to amplify the L. pneumophila pmrA and pmrB genes by PCR were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA), and are as follows: pmrA, forward (TTACTTGTTGAAGACGATGAAC) and reverse (ACGTATAGTGCGGATAAAGTT) and pmrB, forward (CTATTACTACAACATTAACTGCTATT) and reverse (TTTTGGTTTTGTTTTGATGG). Kan insertion was verified by PCR using the manufacturer Kan primers and either the forward or reverse primer for each gene. In both genes, Kan was inserted in the first 300 bp.
Bacterial strains and media.
The virulent strain of L. pneumophila AA100/130b (ATCC BAA-74) is a clinical isolate that has been described previously (2). The isogenic dotA and htrA mutants of strain L. pneumophila AA100 has been described previously (26, 27, 54). Bacteria were grown from frozen stocks on buffered charcoal-yeast extract (BCYE) agar at 37°C or in buffered yeast extract (BYE) broth at 37°C with shaking (20) for 3 days. The plates and broth used for the cultivation of the mutants were supplemented with 50 μg of Kan/ml. E. coli strain DH5α was used as surrogate to clone the pmrA and pmrB genes. The WT L. pneumophila that was used for the confocal rescue studies harbored the plasmid pAM239, which encodes gfp (54). The plates for the gfp-transformed strains were supplemented with 5 μg/ml of chloramphenicol. E. coli strains were cultured with the appropriate antibiotic on Luria-Bertani (LB) agar plates at 37°C with 5% of CO2 or in LB broth at 37°C with shaking.
Assays for survival under stress conditions.
L. pneumophila WT strain AA100 and both pmrA and pmrB mutants were grown for 3 days on BCYE plates at 37°C; 20 ml of BYE medium were then inoculated, and cells were grown for at least 24 h for the stationary-phase stress experiments. The initial CFU count used to inoculate broth for both the E and the PE phases was ∼108 per ml. The absorbance at 550 nm was measured by using a thermospectronic spectrophotometer (Thermo Fisher Scientific, Waltham, MA). For E-phase cultures, an optical density (OD) between 0.4 and 0.8 was used; an OD between 3 and 4 was used for the PE-phase cultures. Cells were centrifuged and resuspended in an equal volume of 1× M63 salts to measure the untreated CFU. M63 salts medium contains 22.0 mM KH2PO4, 40.2 mM K2HPO4, 14.6 mM (NH4)2SO4, and 500 nM FeSO4 (pH 6.5). For the different stress conditions, the cell pellet was resuspended in an equal volume solution of 5 M sodium chloride or 0.1 M citric acid at pH 3 for acid stress. Cells were incubated in a 37°C water bath for 30 min. The cells were washed with 1× M63 salts and serially diluted to determine the CFU on BCYE agar plates. To measure the sodium sensitivity of both the WT strain and the mutants, BYE cultures grown to E and PE phases were diluted into H2O and then plated on BCYE that did or did not contain 100 mM NaCl. The percentage of bacteria that were sodium resistant was calculated as described previously (11).
Microarray analysis.
Gene expression analyses of three independent overnight axenic cultures of each of the mutant strains in both E and PE growth phases were performed. The parental strain AA100 and each of the isogenic mutants (pmrA and pmrB) were inoculated into 50 ml of BYE in 250-ml baffled flasks at an OD at 600 nm (OD600) of 0.05 and grown at 37°C in a rotary shaker at 250 rpm. Each of the cultures was sampled in two 2-ml aliquots at the mid-exponential growth phase (OD600 ∼ 0.8) and then at the PE growth phase (2 to 3 h upon cessation of growth, OD600 = 3). The bacteria in the collected samples were pelleted by centrifugation and stored frozen at −80°C. The total RNA was isolated from the bacteria by using Qiagen RNeasy RNA isolation procedure. To prepare the samples for microarray hybridization, 20 μg of total RNA from each of the samples was converted to cDNA by reverse transcription in the presence of allylamino-dUTP and fluorescently labeled by coupling the resulting cDNA with the fluorescent dyes Alexa Fluor 546 or Alexa Fluor 647 (Invitrogen) according to the manufacturer's recommendations. The whole-genome gene expression profiling of the mutant strains was done using a custom-made longmer (70-mer) oligonucleotide array designed and manufactured at Columbia Genome Center (Columbia University). A set of 2,977 longmer oligonucleotide probes corresponding to all unique genes identified in the L. pneumophila strain Philadelphia-1 was prepared by using the custom oligonucleotide synthesis service from MWG Biotech, Inc. (High Point, NC). The longmer probes were dissolved in 50% dimethyl sulfoxide at 30 μM and spotted in duplicates onto Corning UltraGAPS-coated slides (Corning, Inc., MA) using SpotArray 72 spotting robot (Perkin-Elmer). After the spotting, the microarray slides were stored desiccated at room temperature until further use. The downstream processing and hybridization of the microarrays was performed according to the protocols recommended by Corning, Inc.
After hybridization, the arrays were scanned by using ScanArray Express (Perkin-Elmer, MA) at 5-μm resolution, and the resulting hybridization intensities for all probes from both channels on each array were exported in tab-delimited text file format for further analysis. Raw signal intensities were corrected for dye labeling effects within and between all slides by using the cyclic Lowess procedure implemented in the bioconductor affy microarray analysis package (30). Statistically significant differential expression between the mutants and the AA100 strain at different growth phases was determined by using the unpaired two-tailed t test statistics implemented in the SAM package (72). False discovery rates for the datasets were subsequently estimated as q-values (69). The delta parameters for the q-value cutoffs were set, allowing less than 5% median false positives in a data set. The resulting data were imported for filtering and visualization into the Spotfire DecisionSite for Functional Genomics software suite (TIBCO Spotfire, Inc.). The results were filtered for ScanArray quality scores greater than 2. The resulting sets of differentially expressed genes were further analyzed by using principal component analysis and hierarchical clustering algorithms implemented within Spotfire DecisionSite for Functional Genomics.
Real-time quantitative reverse transcriptase PCR.
For analysis of expression of the flaA gene in vitro, samples of bacterial cultures from the WT strain AA100 and the pmrA and pmrB mutants were grown in BYE medium to an OD550 of 0.8 to 1 (E phase) or an OD550 of 2.0 to 2.2 (PE phase). Total RNA was extracted by using RNeasy minikit (Qiagen) as recommended by the manufacturer. RNA integrity was assessed by visualizing ethidium bromide-stained 0.8% agarose gel. Total RNA was treated with DNase I (Ambion, Austin, TX) at 37°C for 30 min. Equal amounts of RNA were used for cDNA synthesis with Superscript III Plus RNase H− reverse transcriptase (Invitrogen) and random primers. The generated cDNA was diluted fivefold with RNase-free water. Real-time quantitative PCR was done by using the DNA Engine Opticon System (MJ Research) and carried out in triplicates using a DyNAmo Sybr green quantitative PCR kit in a 20-μl reaction volume, as recommended by the manufacturer (New England Biolabs, Ipswich, MA). The 16S RNA and flaA were amplified using primers described previously (32). The 16S RNA was used as an internal normalizing control and to confirm that an equal amount of total RNA was used in each reaction. The PCR conditions were 5 min at 94°C, 15 s at 96°C, and 15 s at 72°C for 30 cycles. The concentration was determined by using the comparative cycle threshold (CT) method (i.e., the CT number at the cross-point between the amplification plot and the threshold) and normalized values to the 16S RNA. Relative quantitation by quantitative reverse transcriptase PCR was validated by equivalent and linear amplification of 16S RNA and the flaA gene at the assay concentrations. Negative or positive values were considered downregulation or upregulation of flaA gene expression, respectively, as represented by a minimum twofold difference.
Cell cultures.
Isolation and preparation of the human monocyte-derived macrophages (hMDMs) and macrophage-like U937 cells was carried out as previously described (60). Cells were maintained in RPMI 1640 tissue culture medium (Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum fetal bovine serum (Gibco-BRL). Axenic Acanthamoeba polyphaga was cultured as adherent cells in PYG medium as previously described (27). Tetrahymena pyriformis was grown in plate count broth (Becton Dickinson Microbiology Systems) (5). With the exception of the growth of T. pyriformis at 25°C, all cells were grown at 37°C in the presence of 5% of CO2.
Intracellular growth kinetics.
The bacterial strains were grown in BYE medium to an OD550 of 2.0 to 2.2 (PE phase). The mammalian or protozoan cells were infected with the bacteria (27) at a multiplicity of infection (MOI) of 10, but an MOI of 100 was used for T. pyriformis. To synchronize the infection, the plates were centrifuged for 5 min at 1,000 rpm using a Centra GP8R Thermo IEC centrifuge. After 1 h of incubation in 5% CO2 at 37°C, the infected cells were washed three times with the culture medium to remove extracellular bacteria and incubated with 50 μg of gentamicin/ml for 1 h to kill the remaining extracellular bacteria. This step was considered the zero time point (t0), and the infected cells were subsequently incubated for several time intervals. At the end of each time interval, the culture supernatant was removed, and the macrophages were lysed hypotonically by the addition of 200 μl of sterile water for 10 min or with 0.04% Triton X-100 for the protozoan cells. The supernatant and the lysates were combined, serial dilutions were prepared, and aliquots were plated on BCYE plates for counting. The number of bacteria was expressed as the number of CFU/ml.
CLSM.
Confocal laser scanning microscopy (CLSM) and sample analysis were performed with polyclonal rabbit anti-L. pneumophila antiserum and Alexa Fluor 488-conjugated donkey anti-rabbit immunoglobulin G (IgG) purchased from Invitrogen. The anti-LAMP-2 (H4B4) monoclonal antibody (developed by J. T. August and J. E. K. Hildreth) was obtained from the Developmental Studies Hybridoma Bank (University of Iowa). To label the lysosomes, monoclonal anti-cathepsin D antibody (BD Transduction, Franklin Lakes, NJ) was used. Mouse anti-KDEL monoclonal antibody purchased from StressGen Biotechnologies (Ann Arbor, MI) was used to label the ER proteins, followed by the addition of Alexa Red 555-conjugated donkey anti-mouse IgG (Molecular Probes). To study the role of PmrA and PmrB in intracellular replication, approximately 5 × 105 hMDMs were grown on circular glass coverslips (Fisher Scientific, Pittsburgh, PA) in 24-well culture plates. An MOI of 10 was used for all experiments for the CLSM except for when formalin-killed bacteria was used as a control, when an MOI of 50 was used. After infections, cells were washed three times with phosphate-buffered saline and processed for confocal microscopy as described previously (60).
For the coinfection experiments, cells were coinfected simultaneously with the WT strain of L. pneumophila expressing green fluorescent protein (GFP) and isogenic mutants at an MOI of 10, with the exception of dotA mutant, for which an MOI of 20 was used. To synchronize the infection, the plates were centrifuged for 5 min at 1,000 rpm using a Centra GP8R Thermo IEC centrifuge. After 1 h of incubation in CO2 at 37°C, the infected cells were washed three times with the culture medium to remove extracellular bacteria and then incubated with 50 μg of gentamicin/ml for 1 h to kill the remaining extracellular bacteria. Infected cells were further incubated for 10 h, and the cells were processed for confocal microscopy as described below. All bacteria were labeled with polyclonal rabbit anti-L. pneumophila antiserum and Alexa Fluor 555-conjugated donkey anti-rabbit IgG antibody; therefore, the GFP-expressing bacteria become yellow when the two colors are combined, whereas the bacterial strain that did not have GFP is detected by the red fluorescence. The dotA and htrA mutants were used as positive and negative controls, respectively. The cells were examined by using an Olympus Fv500 laser scanning confocal microscope as described previously (60). On average, 8 to 15 0.2-μm serial Z sections of each image were captured and stored for further analyses using Adobe Photoshop 6.0.
TEM.
For transmission electron microscopy (TEM), monolayers in six-well plates were infected with L. pneumophila strains at an MOI of 10 for 1 h, followed by 1 h of gentamicin treatment. At 6 h postinfection, the infected monolayers were washed, and the cells were fixed in 3.5% glutaraldehyde, dehydrated in alcohol, processed, and stained for TEM as we described previously (1). Sections were examined with a Hitachi H-7000/STEM electron microscope at 80 kV (26, 50).
Statistical analysis.
All experiments were performed at least three times, and the data shown are representative of one experiment. To analyze for statistical significant differences between different sets of data, a two-tailed Student t test was used, and the P value was obtained.
Microarray data accession number.
The processed and raw microarray data for gene expression analysis can be obtained from the NCBI Gene Expression Omnibus database. The accession number is GSE13323. Additional descriptions of the microarray platform and the analysis are posted at http://legionella.cu-genome.org/index.html.
RESULTS
L. pneumophila PmrA/PmrB homologs.
We aligned the PmrA response regulator (lpg1292) and PmrB (lpg1291) sensor kinase proteins of L. pneumophila, Coxiella burnetii (Q83CA1 and Q83CA0), Salmonella enterica serovar Typhimurium (AAV92795 and AAA72366), and Pseudomonas aeruginosa (Q88F73 and Q88F74) by using CLUSTAL W software. The alignment results showed that L. pneumophila PmrA shares 60.8, 45.3, and 55.1% identity and 75.8, 63.6, and 70.7% similarity with C. burnetii, S. enterica serovar Typhimurium, and P. aeruginosa, respectively. PmrA alignments were also performed in other bacteria, including E. coli, Erwinia carotovora (Q6DB90 and Q6DB91), and Chromobacterium violaceum (Q7NZN1 and Q7NZN2). These results showed an average of 85.2% of consensus sequence was shared in all of them (data not shown).
The PmrB sensor shares 47.8, 23.95, and 40.1% identity and 60.7, 38.6, and 60% similarity to C. burnetii, S. enterica serovar Typhimurium, and P. aeruginosa, respectively. Using the SOSUI system (http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html), the predicted topology of the PmrB protein in C. burnettii, S. enterica serovar Typhimurium, and P. aeruginosa indicated one to two periplasmic domain(s) where the C-terminal region exposed to the outside harbors the sensor domain that becomes activated by a specific environmental signal. Alignment of the PmrB sensor protein sequences of L. pneumophila, C. burnetii, and serovar Typhimurium revealed the presence of an EXXE consensus sequence surrounded by a basic amino acid, which is a lysine in L. pneumophila. This EXXE motif was shown to be present in the Saccharomyces FTR1 iron transporter and in the mammalian ferritin light chain (78), suggesting that iron might be a signal recognized by the PmrB sensor.
Phenotypic characteristics of pmrA and pmrB mutants under different stress conditions.
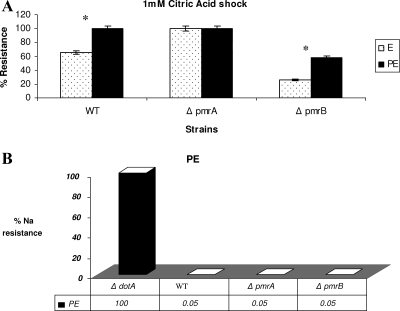
In S. enterica serovar Typhimurium, the sensor kinase PmrB was identified as the primary sensor that activates the PmrA protein when the pathogen experiences strong acid pH, resulting in the transcription of PmrA-activated genes (55). To determine whether low pH may trigger the pmrAB TCS in L. pneumophila, we exposed both the pmrA and the pmrB mutants grown to E and PE phases to 1 mM citric acid (pH 3). At 30 min after exposure, the pmrB mutant showed a significant growth defect compared to the WT strain and the pmrA mutant (Fig. 1A, P < 0.1). The complemented pmrB mutant was similar to the WT in sensitivity to acidic pH (data not shown).
FIG. 1.
Phenotypic characteristics of the pmrA and pmrB mutants under different stress conditions. For the different stress conditions, the cell pellet was resuspended in an equal volume solution of 0.1 M citric acid at pH 3 for acid stress (A) or 5 M sodium chloride (B) and compared to bacteria resuspended in M63 salts. Cells were incubated in a 37°C water bath for 30 min and then washed, serially diluted, and plated on BCYE agar plates to determine the number of resistant bacteria. The experiment was done three times, and the data are representative of one independent experiment. Asterisks represent a significant difference between the WT strain and the pmrB mutant.
It was shown that the PE-phase WT strain L. pneumophila is sodium sensitive and osmotically resistant (11). However, mutants such as the dot/icm mutants of L. pneumophila that have lost sensitivity to sodium ion are salt resistant (10, 62). We show here that the pmrA and pmrB mutants were salt sensitive to the same extent as the WT strain (Fig. 1B). Moreover, when subjected to osmotic stress (5 M NaCl), L. pneumophila WT strain and the pmrA and pmrB mutants showed no significant difference in osmotic resistance at the E or PE phases (data not shown).
Microarray gene analysis of the pmrA and pmrB mutants.
The PmrA response regulator has been recently shown to control the expression of 13 tested substrates exported by the Dot/Icm secretion system (79). The role of this regulator in global regulation of the L. pneumophila genome, as well as its transcription profile, is not known. To address this, genome-wide microarray analyses were performed, and gene expression in both the pmrA and the pmrB mutants was compared to the WT strain AA100 at both the E and the PE phases (see Table S1.1 in the supplemental material). Keeping in mind that the pmrAB locus is present in all L. pneumophila sequenced genomes and that gene variation among L. pneumophila strains usually represents a small proportion of the genome (53), we performed microarray analysis using a Philadelphia-1 strain-specific microarray. A total of 279 genes in both the pmrA and the pmrB mutants were differentially expressed compared to the WT strain (the findings are summarized in Table 1). The genes were divided into nine groups as follows: (i) ceg genes or genes with the consensus sequence for the PmrA binding (32 genes were downregulated in both pmrA and pmrB mutants, and four genes were upregulated in the pmrB mutant at the PE phase); (ii) eukaryotic-like protein encoding genes (10 were downregulated in both mutants during both growth phases, and 3 were upregulated mainly at the PE phase); (iii) type IV secretion apparatus encoding genes and Dot/Icm translocated-effectors (26 genes were downregulated in both mutants at the E and PE phases, and 3 were upregulated at the PE phase); (iv) substrates secreted by the type II secretion system (7 genes show downregulation in both mutants, and only 2 were upregulated in either mutant at the PE phase); (v) stress response genes (13 genes were downregulated mainly at the E phase in the pmrA mutant, and 11 were upregulated in both mutants mainly at the PE phase); (vi) PE-phase regulators (2 genes were downregulated at both phases in both the pmrA and the pmrB mutants); (vii) flagellar genes and other operons (18 genes were downregulated and 15 were upregulated in both mutants at both growth phases; all flagellar synthesis genes [flgB to flgL] were upregulated in both mutants at the PE phase); (viii) genes encoding for proteins involved in various metabolic pathways (14 genes were upregulated in the mutants, and 4 were downregulated mainly at the PE, in the pmrB mutant only); and (ix) 127 genes of unknown function (see summary in Table 1).
TABLE 1.
Summary of genes regulated in a PmrA/PmrB-dependent manner at the E and PE phases
| Regulation type | Cegb | No. of genesa
|
Total no. of genes | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Eukaryotic-like | Type IV secretion system | Type II secretion system | Stress response | PE-phase regulators | Flagellar genes, other operons | Metabolic genes | Unknown function | |||
| Positively regulated | 32 | 10 | 26 | 7 | 13 | 2 | 18 | 14 | 120 | 242 |
| Negatively regulated | 4 | 3 | 3 | 2 | 11 | 0 | 15 | 4 | 7 | 49 |
| Total no. | 36 | 13 | 29 | 9 | 21* | 2 | 24* | 18 | 127 | 279* |
Totals indicated by an asterisk exclude genes that were inversely regulated at the E and PE phases.
Ceg, no. of genes coregulated with the effector encoding gene.
Our data showed that only 36 ceg genes were differentially expressed in a PmrA/PmrB-dependent mechanism (see Table S1.2 in the supplemental material). A large number of genes encoding eukaryotic-like proteins, including some ankyrin genes (lpg0038 and lpg2452), a zinc metalloproteinase, and a uracyl DNA glycosylase, were positively regulated by PmrA at both the E and the PE phases (see Table S1.3 in the supplemental material). Upregulation of the expression of ankyrin genes by PmrA and PmrB as derived from the microarrays was confirmed by real-time PCR analyses (data not shown). Our data also showed that the expression of 7 dot/icm apparatus encoding genes (dotA, icmL, icmR, icmV, icmW, and icmX), as well as 22 Dot/Icm-secreted effector encoding genes, was upregulated in both pmrA and pmrB mutants at both growth phases. This group includes sde-like genes (lpg2153 and lpg2154) that are considered virulence factors of the transmissive phase (44) and most of the sid-related genes (sidC, sidD, sidE, and sidF). The exceptions were sidA and sidG, both of which were downregulated in a PmrA-dependent manner. The positive regulation of PmrA over sidH (lpg2829) was observed in the PE phase only (see Table S1.4 in the supplemental material). Interestingly, PmrA was found to regulate eight type II-secreted effectors, including Acph-1, icmX, lvrE, dnaK, zinc metalloprotease, and chitinase encoding genes and two genes of unknown functions (lpg2526 and lpg1385) (see Table S1.5 in the supplemental material) (18). Therefore, PmrA is a major transcriptional regulator of genes encoding substrates exported by the type IV and type II secretion systems.
Interestingly, the type IV pilin gene, pilE, involved in L. pneumophila adherence to mammalian and protozoan cells was downregulated in the pmrB mutant at the PE phase only (66). Also, three of the Legionella vir homologues (lvh)—lvrE, virB11, and virD4—were downregulated in the pmrB mutant at the PE phase.
Fourteen genes encoding metabolic enzymes (aroE, phbC, maeA, and bdhA) were upregulated in a PmrA/PmrB-dependent manner, mainly at the E phase of growth, suggesting that L. pneumophila may be using this TCS to couple its differentiation to the metabolic state (see Table S1.9 in the supplemental material). The expression of 20 genes encoding for chaperones and heat and cold shock proteins was downregulated in the pmrA mutant in the exponential phase, but this suppression was alleviated at the PE phase. Some of the chaperons genes, including hslVU protease subunits (lpg0640 and lpg0641), hsp10 and hsp60 (lpg0687 and lpg688), and dnaK and grpE (lpg2025 and lpg2026), were grouped into operons or located adjacent to each other (see Table S1.6 in the supplemental material).
Two major PE-phase regulators, the CsrA activator of replication and repressor of transmission traits of L. pneumophila, as well as the RpoE sigma factor encoding genes (lpg0781 and lpg1577), was regulated in a PmrA/PmrB-dependent manner. The global regulator encoding gene csrA was downregulated in both mutants at both growth phases, whereas the rpoE was downregulated in both mutants only at the PE phase of growth (see Table S1.7 in the supplemental material).
In addition, several operons controlling the expression of flagellar basal body (flgB to flgL) and located in the same 10-kb region of the Legionella genome spanning loci lpg1216 to lpg1226 are upregulated in the pmrA and pmrB mutants during the PE phase only (see Table S1.8 in the supplemental material). To confirm the role of the PmrA/PmrB TCS in the expression and regulation of the flagellar genes, we measured the expression levels of the flaA gene by real-time PCR. The flagellum subunit protein encoding gene flaA was increased by ninefold in the pmrA mutant at the PE growth phase compared to the WT strain (see Fig. S1 in the supplemental material). Therefore, regulation of flagellar gene expression, rpoE, and the global repressor protein (csrA) indicated that PmrA was a major global regulator of L. pneumophila, particularly at the PE phase. Interestingly, a group of 41 genes did not show the same pattern of regulation in both the pmrA and the pmrB mutants, suggesting the presence of cross talk between the PmrA/PmrB and other TCSs (see Table S2 in the supplemental material).
Role of PmrA and PmrB in intracellular growth of L. pneumophila within amoebas and ciliates.
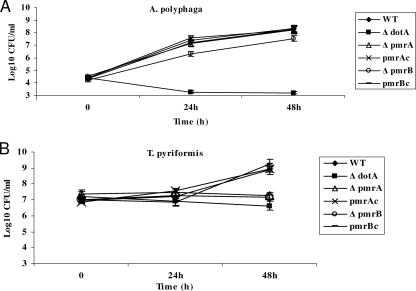
Previous work has shown that a pmrA mutant constructed in the JR32 strain of L. pneumophila is defective for intracellular growth in Acanthamoeba castellanii, but the defect was not restored by the WT genes in trans, suggesting a secondary mutation may have partially resulted in the defect (79). However, the role of PmrB in the intracellular infection remains unknown. We have constructed the pmrA and pmrB mutants in strain AA100/130b and examined their growth in two different protozoan hosts: A. polyphaga and the ciliate T. pyriformis. The growth rate of the pmrA and pmrB mutants in BYE broth showed growth rates similar to that of the WT strain, and there was no difference in the length of the lag phase (data not shown). The pmrA mutant grew normally in A. polyphaga, whereas the pmrB mutant was partially defective, since 10-fold fewer CFU were recovered at both 24 and 48 h (Fig. 2A). In contrast, a more severe growth defect was observed for both mutants in T. pyriformis. At 48 h postinfection, there was no detectable growth for the pmrA and pmrB mutants in the ciliate, and the defect was fully complemented by the WT gene (Fig. 2B). As expected, the dotA mutant control was not able to grow in both hosts. We conclude that both proteins played an important role in intracellular growth within the ciliate T. pyriformis.
FIG. 2.
Intracellular growth kinetics of the pmrA and pmrB mutants of L. pneumophila within protozoa. The intracellular growth kinetics of the pmrA and pmrB mutant in A. polyphaga (A) and T. pyriformis (B) were determined. The infection was carried out in triplicates for 1 h, followed by 1 h of gentamicin treatment to kill extracellular bacteria in case of A. polyphaga. The infected monolayers were lysed at different time intervals and plated onto agar plates for colony enumeration. The experiment was done three times, and the data are representative of one independent experiment. Error bars represent standard deviations, but some were too small to appear in the figure.
Role of PmrA and PmrB in the intracellular growth of L. pneumophila within human macrophages.
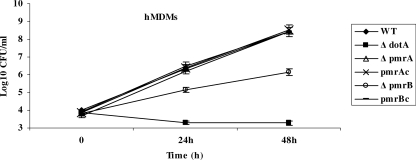
Zusman et al. (79) have previously reported that a pmrA mutant derived from the JR32 L. pneumophila strain is partially defective in the HL-60 macrophage cell line. However, whether PmrB plays any role in the intracellular infection of mammalian macrophages is not known. To elucidate the role of both PmrA and PmrB proteins in the intracellular growth of L. pneumophila in human macrophages, we examined the intracellular growth kinetics of both mutants. We assessed the role of the two proteins in intracellular replication by examinating the intracellular growth kinetics of the mutants within hMDMs and the U937 macrophage cell line (Fig. 3 and data not shown). Our data showed that the pmrA mutant had no detectable intracellular growth defect in both cells. In contrast, the pmrB mutant was defective in hMDMs, with 200-fold fewer CFU recovered after 48 h compared to the WT strain (Fig. 3). In U937 cells, 2,000-fold fewer CFU were recovered for the pmrB mutant compared to the WT strain by 48 h postinfection (data not shown). In both cases, the defect was fully complemented by the WT gene. As expected, the dotA mutant control did not grow within any of the macrophages tested (Fig. 3). We conclude that the intracellular growth phenotype of both mutants in human macrophages is different, which further supports our speculation of a possible cross talk between the PmrA/PmrB and other TCSs.
FIG. 3.
Intracellular growth kinetics of the pmrA and pmrB mutants of L. pneumophila within macrophages. Intracellular growth kinetics of the WT AA100 strain and the dotA, pmrA, and pmrB mutants in hMDMs. pmrAc and pmrBc represent the pmrA and pmrB mutant strains complemented with the WT copy of the gene on the pBC plasmid. The infection was carried out in triplicates for 1 h at an MOI of 10, followed by 1 h of gentamicin treatment to kill the extracellular bacteria. The infected monolayers were hypotonically lysed at the indicated time points after infection and plated onto agar plates for colony enumeration. The data are representative of three independent experiments, and error bars represent the standard deviations.
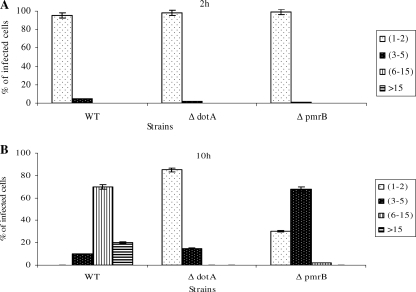
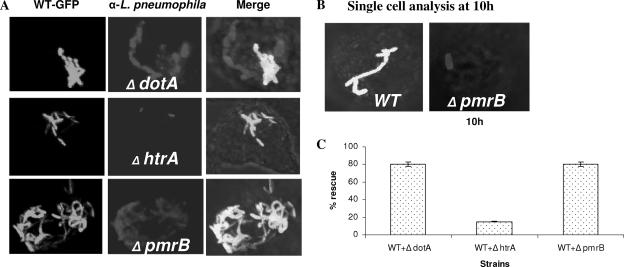
Since the L. pneumophila pmrB mutant exhibited a defect in intracellular replication, we examined whether this defect was due to a defect in replication of a subset of bacterial population or to a balanced killing and replication. We performed single-cell analysis to quantitate the number of bacteria per cell at several stages of the intracellular infection of hMDMs. The data showed that at 2 h after infection, ca. 98% of the cells infected with the different strains harbored one organism (Fig. 4A). After 10 h of infection, ca. 70% of the cells harboring the WT strain contained 6 to 15 bacteria. In contrast, the pmrB mutant showed less replication, with ca. 68% of the cells harboring three to five bacteria per cell (Fig. 4B). We conclude that the PmrB sensor protein plays an important role in the intracellular growth of L. pneumophila within macrophages and that the intracellular growth defect caused by the pmrB mutation is homogeneous.
FIG. 4.
Single cell analyses of replicative phagosomes. At 2 and 10 h postinfection of hMDMs, 100 infected cells were analyzed by CLSM for the formation of replicative phagosomes. Representative quantitation of the number of bacteria/cell at 2 h (A) and 10 h (B) is shown. The dotA mutant was used as a negative control. Infected cells from multiple coverslips were examined in each experiment. The results are representative of three independent experiments performed in triplicates. Error bars represent the standard deviations.
in cis rescue of the pmrB mutant within communal phagosomes harboring the WT strain.
The dot/icm mutants are rescued in cis for their intracellular defect within communal phagosomes harboring the WT strain of L. pneumophila that is able to modulate phagosomal biogenesis into a niche suitable for bacterial replication (17). However, L. pneumophila mutants defective in intracellular replication due to a defect in stress response genes (such as htrA or rpoS), which are required for adaptation to the phagosomal microenvironment, are not rescued in cis within communal phagosomes harboring the WT strain (4, 17, 54). To examine whether the PmrB protein is required for formation of replicative phagosomes or for adaptation to the phagosomal microenvironment, we coinfected hMDMs with the WT strain and the pmrB mutant and determined whether the mutant replicated in communal phagosomes harboring the WT strain. We used coinfection of WT L. pneumophila and the two isogenic mutants, the dotA or htrA mutant, as positive and negative controls, respectively. In all coinfections, only ∼10% of the phagosomes were communal phagosomes harboring the two different strains. In the control coinfection of L. pneumophila and its dotA mutant, replication of the dotA mutant was rescued in communal phagosomes containing the WT strain (Fig. 5A and C). Control coinfection of L. pneumophila and its htrA mutant showed the failure of the WT strain to rescue the htrA mutant in communal phagosomes (Fig. 5A and C) (54). Compared to the replication status of a single infection by the WT strain and the pmrB mutant at 10 h postinfection (Fig. 5B), our data showed that when the pmrB mutant resided in communal phagosomes harboring the WT strain, the mutant replicated robustly, similar to what was observed with the dotA mutant (Fig. 5A and C). We conclude that, despite the severe growth defect caused by the pmrB mutation, the pmrB mutant is able to replicate within a vacuole remodeled by the WT strain similar to the dot/icm structural mutants (17).
FIG. 5.
in cis rescue of intracellular growth of the pmrB mutant within communal phagosomes harboring the WT strain in hMDMs. The hMDMs were simultaneously infected with the GFP-positive L. pneumophila strain AA100 (WT) and one of the mutants—dotA, htrA, or pmrB—followed by fixation at 10 h after infection (see Materials and Methods). Macrophages harboring phagosomes containing both strains (GFP-WT and the mutants) were scored. Representative confocal images and quantitation are shown in panels A and C, respectively. Panel B shows the replication status of single infection by the WT strain and the pmrB mutant at 10 h postinfection. The results are representative of three independent experiments performed in triplicates. Error bars represent the standard deviation.
Intracellular trafficking of the pmrB mutant within hMDMs.
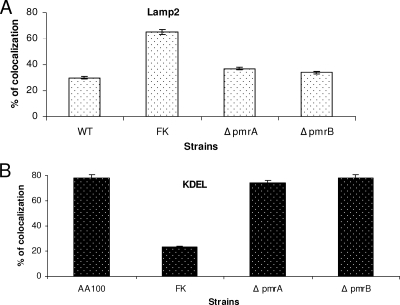
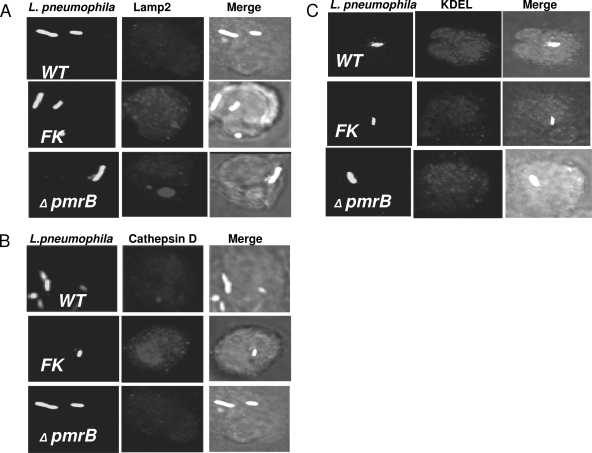
Since the pmrB mutant was defective in intracellular replication, we examined whether its growth defect was caused by a failure to evade the endocytic pathway. To examine the intracellular trafficking of the pmrB mutant within hMDMs, we labeled the cells with the late endosomal/lysosomal marker (LAMP-2) and the luminal lysosomal enzyme cathepsin D. CLSM was used to assess the percentage of colocalization of phagosomes harboring the WT strain AA100 or the pmrB mutant with the late endosomal and lysosomal compartments (Fig. 6 and 7). Formalin-killed bacteria, which traffic to the phagolysosomes (41, 42), were used as a positive control. The data showed that at 2 h postinfection, phagosomes harboring the WT strain and the pmrB mutant colocalized with LAMP-2 at levels of 30 and 34%, respectively, whereas phagosomes containing formalin-killed bacteria showed 65% colocalization (Fig. 6A and 7A). Approximately 62% of the phagosomes harboring formalin killed bacteria colocalized with the lysosomal marker cathepsin D, whereas the WT strain AA100 and the pmrB mutant showed 31 to 40% colocalization, respectively (data not shown, Fig. 7B). The slight difference in Lamp2 and cathepsin D colocalization between the WT strain and the pmrB mutant was not significant (Student t test, P > 0.1). The results were similar for the pmrA mutant trafficking (Fig. 6). We conclude that, despite its role in intracellular growth, PmrB is not involved in the regulation of genes required for evasion of the endocytic pathway.
FIG. 6.
Quantitative analysis of intracellular trafficking of the pmrA and pmrB mutants within hMDMs. Quantitation of infected hMDMs for colocalization of the bacterial phagosome for the WT strain AA100 and the pmrA and pmrB mutants with the late endosomal marker LAMP-2 (A) at 2 h postinfection and the ER marker KDEL at 4 h (B) was performed. Formalin-killed (FK) bacteria were used as a negative control. At least 100 infected cells from multiple coverslips were examined in each experiment by CLSM. The results shown are representative of three independent experiments performed in triplicates. The data represent means ± the standard deviation. There was no significant difference in trafficking of the WT strain and the mutants.
FIG. 7.
Intracellular trafficking of the pmrA and pmrB mutants of L. pneumophila within hMDMs. Representative confocal microscopy images of infected hMDMs show colocalization of the bacterial phagosome with the late endosomal marker LAMP-2 (A), the lysosomal enzyme cathepsin D (B), and the ER marker KDEL (C). The bacteria and the LAMP-2, cathepsin D, and KDEL markers were detected by specific antibodies. Formalin-killed (FK) bacteria were used as a negative control. The results shown are representative of three independent experiments performed in triplicates.
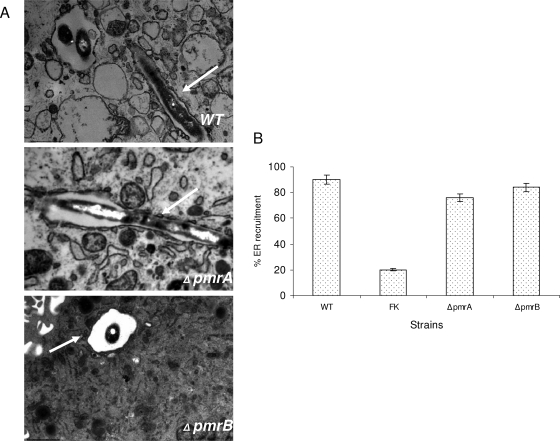
Using confocal microscopy, we determined the capacity of the pmrA and pmrB mutants to decorate their vacuoles with the ER-derived vesicles using an antibody that recognizes the KDEL amino acid sequence, which is the signal for ER retention. The data showed that at 4 h postinfection of hMDMs, 70 to 80% of the phagosomes harboring the WT strain and the pmrA and pmrB mutants colocalized with the KDEL marker. The formalin-killed bacteria used as a negative control were defective in acquiring the KDEL marker, where only 23% of these phagosomes retained the KDEL maker (Fig. 6B and 7C). These results were consistent with the TEM findings, where we examined the rough endoplasmic reticulum (RER) recruitment to the phagosomes at 6 h postinfection. No significant difference (Student t test, P > 0.1) was observed between the WT strain (90%) and the pmrB mutant (84%) containing vacuoles (Fig. 8). These data indicate that the PmrA/PmrB TCS is not involved in the regulation of genes required for ER recruitment to the phagosome.
FIG. 8.
The L. pneumophila PmrA/PmrB mutants are contained within ER-derived phagosomes. (A) Cells were examined by TEM for the presence of the RER studded phagosome at 6 h. The ER is indicated by arrows in the representative electron micrographs shown. (B) Quantitative results for the pmrA and pmrB mutants compared to the WT strain AA100 and the formalin-killed (FK) WT strain. The results are expressed as percentage of 100 phagosomes surrounded by the RER. The experiment was done three times in triplicate, and error bars represent the standard deviations. There was no significant difference in trafficking of the WT strain and the mutants.
DISCUSSION
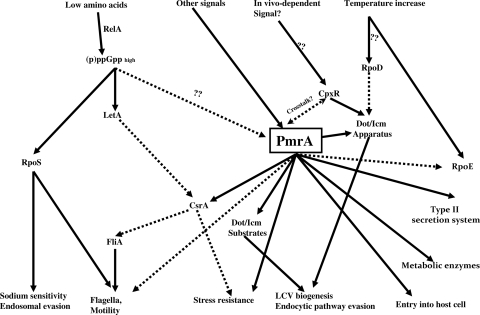
Expression of L. pneumophila virulence factors has been proposed to be mediated by a regulatory cascade that is triggered upon nutrient starvation and driven via two parallel pathways: one involving LetA/LetS and the other involving the RpoS sigma factor (Fig. 9) (8, 9, 47). Recently, the PmrA/PmrB TCS has been shown to control the expression of 13 tested L. pneumophila genes encoding Dot/Icm-secreted effectors (79). However, the role of this TCS in the regulation of other virulence factors, as well as its implication in the regulatory cascade of L. pneumophila that governs phenotypic transition at the PE phase, is not known.
FIG. 9.
Working model of the regulatory cascade governing the life cycle phenotypic switch of L. pneumophila. Solid lines indicate positive regulation. Dashed lines indicate negative regulation (see Discussion for details).
Based on the rationale that genes present in all four sequenced L. pneumophila strains represent the core genome of this species (14, 15), we performed microarray analysis using a Philadelphia-1 strain-specific microarray. The four sequenced genomes of L. pneumophila contain each an average of 3,000 genes with 200 to 300 genes specific to each genome, but all contain the pmrAB locus. It is important to note that some of the genes present in the clinical strain AA100/130b may be absent in the Philadelphia-1 strain and vice versa and that gene variation among bacterial strains of the same species usually represents a small proportion of the genome (53). Despite potential minor differences between the Philadelphia-1 strain genome and our clinical strain AA100/a30b, ∼10% of the core genome appears to be under the regulation of PmrA.
Using a genome-wide microarray, we have shown for the first time that the PmrA response regulator not only regulates the expression of some Dot/Icm secreted effectors but is a global regulator of L. pneumophila. The PmrA/PmrB TCS controls the expression of 279 genes of L. pneumophila. Although PmrA in E. coli, P. aeruginosa, and S. enterica serovar Typhi has been shown to activate genes involved in the modification of lipopolysaccharide and subsequent resistance to antimicrobial peptides (31), our microarray data do not show any regulation of lipopolysaccharide modification genes by the PmrA/PmrB regulon of L. pneumophila.
We propose a working model of the global role of PmrA/PmrB in regulation (Fig. 9). The accumulation of ppGpp stimulates both RpoS and the LetA/LetS cascades of regulation (28, 33, 38). Both cascades result in the expression of many of the virulence traits, causing a major shift in bacterial differentiation from the replicative phase to the transmissive phase (28, 33, 38). This ppGpp alarmone may also induce the PmrA/PmrB cascade, since PmrA expression is induced further at the PE phase of growth (data not shown). Upon expression and/or activation of the PmrA by the PmrB sensor, PmrA modulates the expression of genes that account for ∼10% of the core genome. We show that flagellar genes are downregulated in PmrA-dependent manner, which may be acting through that activation of the CsrA repressor. Regulators of the PE phase, LetA/LetS and RpoS, positively regulate flagellar genes upon entry into the PE phase of growth (9, 47). We show here that PmrA is the first negative regulator of expression of flagellar genes at the PE phase. Despite the downregulation of flagellar expression, both the pmrA and the pmrB mutants were motile (data not shown). Therefore, the role of other regulators, such as RpoS, in inducing flagellar genes at the PE phase of growth tips the balance in favor of flagellar gene expression. Therefore, PmrA/PmrB may act as a negative-feedback loop to fine-tune flagellar gene expression.
We show that metabolic enzymes (PhbC, MaeA, HydG, BdhA, and AroE) are regulated in a PmrA/PmrB-dependent mechanism, suggesting that PmrA/PmrB is involved in certain aspects of the transition of L. pneumophila from the E phase to the PE phase upon nutrient starvation (61). Therefore, PmrA-dependent regulation may act as a link between nutrient acquisition and microbial differentiation. Our data clearly show that PmrA acts as a global regulator of genes involved in the transition from the replicative to the transmissive phase of growth and modulates a variety of L. pneumophila cellular, metabolic, and physiological processes, particularly at the PE phase. None of the PE-phase regulators RelA, LetA/S, and RpoS appears to be under the direct regulation of PmrA/PmrB TCS. This suggests that PmrA may be downstream of these regulators or that PmrA is acting through a different regulatory cascade. We speculate that by coupling differentiation to the metabolic state through the PmrA/PmrB TCS, L. pneumophila can swiftly acclimate to environmental fluctuations and stress encountered within or outside the host.
A JR32 strain-derived pmrA mutant of L. pneumophila has been reported to be completely defective for intracellular growth in A. castellanii and partially defective for intracellular growth in HL-60-derived human macrophages, but the introduction of a plasmid containing the pmrA gene has resulted in only partial complementation of the intracellular growth defect within A. castellanii (79). Our data show no detectable defect of the pmrA mutant in both A. polyphaga and human macrophages, but the pmrA mutant is totally defective in the ciliate T. pyriformis. The different phenotype of the pmrA mutant derived from different parental strain (JR32 and AA100) may be due to the different genetic background of the WT strain and/or a difference in the host cells, but the failure of transcomplementation of the JR32 pmrA mutant may indicate a secondary mutation that affected the intracellular growth. The role of PmrB in the intracellular infection is not known. Our data have shown that the pmrB mutant exhibits an intracellular growth defect in human macrophages and A. polyphaga, but the defect is more pronounced for the pmrB mutant within the ciliate T. pyriformis. This suggests a possible role for PmrA and PmrB in conferring protozoan host tropism by L. pneumophila and enabling the bacteria to adapt to different microenvironmental conditions that may vary with different encountered hosts or ecological niches, such as biofilms.
Despite the severe intracellular growth defect within hMDMs, we show that the pmrB mutant evades the endocytic pathway and remodels its phagosome into an ER-derived vacuole. Furthermore, the intracellular growth defect of the pmrB mutant is rescued in cis within communal phagosomes established by the parental strain similar to the dot/icm structural mutants. In contrast, a mutant defective in stress response, such as the htrA mutant, is not rescued within the communal vacuoles established by the WT strain. Our data suggest a role for the PmrA/PmrB TCS in controlling genes involved in bacterial adaptation to the phagosomal microenvironment but not genes required for interception of ER-derived vesicles or evasion of the lysosomes (35).
Our data suggest that PmrA may function through another TCS sensor since the microarray results revealed 41 genes that are differentially regulated in the pmrA and pmrB mutants, and a dramatic difference in phenotypes of the pmrA and pmrB mutants is observed in macrophages as well as in protozoa. Interestingly, a cross talk between the PhoPQ and the PmrA/PmrB TCS has been shown in S. enterica serovar Typhi, where, in addition to the PmrB sensor, PmrA is activated via PmrD, which is a PhoPQ-regulated protein (55). Even though no homologs of PmrD and PhoPQ TCS are present in L. pneumophila, we speculate a possible cross talk with other TCSs such as CpxRA, since some of the PmrA/PmrB-regulated genes have been shown to be regulated by CpxR (icmR, icmV, and icmW) (24).
In summary, we show that the PmrA/PmrB TCS is involved in the intracellular growth of L. pneumophila, enabling the bacteria to adapt to environmental fluctuations through triggering of the PmrA global regulator. This regulator triggers the expression of at least nine large families of genes that are involved in various aspects of L. pneumophila pathogenesis and modulation of cellular processes in macrophages and protozoa. We propose a cross talk between the PmrA/PmrB TCS and other TCSs. The pmrB mutant is defective in mammalian and protozoan hosts, whereas the pmrA mutant shows intracellular growth defect only within ciliates, suggesting that PmrA/PmrB may confer a host tropism to L. pneumophila. Despite the intracellular growth defect of the pmrB mutant, the WT strain rescues its growth defect when both coinhabit the phagosome. Although defective in intracellular proliferation, the pmrB mutant evades the endosomal-lysosomal fusion. We show that the PmrA/PmrB TCS is the first global regulator known in L. pneumophila to negatively regulate flagellar genes, creating a feedback loop of some positive regulators of flagellar expression such as RpoS and LetA/S, which is likely to fine-tune flagellar expression.
Supplementary Material
Acknowledgments
We are in debt to Howard Shuman at Columbia University for the microarray analyses that were supported by his NIH grant RO1 (AI064481).
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 20 October 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abu Kwaik, Y. 1998. Induced expression of the Legionella pneumophila gene encoding a 20-kilodalton protein during intracellular infection. Infect. Immun. 66203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu Kwaik, Y., B. I. Eisenstein, and N. C. Engleberg. 1993. Phenotypic modulation by Legionella pneumophila upon infection of macrophages. Infect. Immun. 611320-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Kwaik, Y., L. Y. Gao, B. J. Stone, C. Venkataraman, and O. S. Harb. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 643127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abu-Zant, A., R. Asare, J. E. Graham, and Y. Abu Kwaik. 2006. Role for RpoS but not RelA of Legionella pneumophila in modulation of phagosome biogenesis and adaptation to the phagosomal microenvironment. Infect. Immun. 743021-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu-Zant, A., M. Santic, M. Molmeret, S. Jones, J. Helbig, and Y. Abu Kwaik. 2005. Incomplete activation of macrophage apoptosis during intracellular replication of Legionella pneumophila. Infect. Immun. 735339-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Khodor, S., C. T. Price, F. Habyarimana, A. Kalia, and Y. Abu Kwaik. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol. Microbiol. 70908-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anand, C. M., A. R. Skinner, A. Malic, and J. B. Kurtz. 1983. Interaction between L. pneumophila and a free-living amoeba (Acanthamoeba palestinensis). J. Hyg. 91167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachman, M. A., and M. S. Swanson. 2004. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect. Immun. 722468-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachman, M. A., and M. S. Swanson. 2001. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol. Microbiol. 401201-1214. [DOI] [PubMed] [Google Scholar]
- 10.Bandyopadhyay, P., B. Byrne, Y. Chan, M. S. Swanson, and H. M. Steinman. 2003. Legionella pneumophila catalase-peroxidases are required for proper trafficking and growth in primary macrophages. Infect. Immun. 714526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne, B., and M. S. Swanson. 1998. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect. Immun. 663029-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cazalet, C., and C. Buchrieser. 2005. What do we learn from the genome of Legionella pneumophila? Med. Sci. 21455-457. (In French.) [DOI] [PubMed] [Google Scholar]
- 13.Cazalet, C., S. Jarraud, Y. Ghavi-Helm, F. Kunst, P. Glaser, J. Etienne, and C. Buchrieser. 2008. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Res. 18431-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 361165-1173. [DOI] [PubMed] [Google Scholar]
- 15.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 3051966-1968. [DOI] [PubMed] [Google Scholar]
- 16.Clemens, D. L., and M. A. Horwitz. 1995. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagolysosomal maturation is inhibited. J. Exp. Med. 181257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coers, J., C. Monahan, and C. R. Roy. 1999. Modulation of phagosome biogenesis by Legionella pneumophila creates an organelle permissive for intracellular growth. Nat. Cell Biol. 1451-453. [DOI] [PubMed] [Google Scholar]
- 18.DebRoy, S., J. Dao, M. Soderberg, O. Rossier, and N. P. Cianciotto. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. USA 10319146-19151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derre, I., and R. R. Isberg. 2004. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect. Immun. 723048-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldman, M., and G. Segal. 2007. A pair of highly conserved two-component systems participates in the regulation of the hypervariable FIR proteins in different Legionella species. J. Bacteriol. 1893382-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 1744317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foussard, M., S. Cabantous, J. Pedelacq, V. Guillet, S. Tranier, L. Mourey, C. Birck, and J. Samama. 2001. The molecular puzzle of two-component signaling cascades. Microbes Infect. 3417-424. [DOI] [PubMed] [Google Scholar]
- 24.Gal-Mor, O., and G. Segal. 2003. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb. Pathog. 34187-194. [DOI] [PubMed] [Google Scholar]
- 25.Gal-Mor, O., T. Zusman, and G. Segal. 2002. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J. Bacteriol. 1843823-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao, L.-Y., O. S. Harb, and Y. Abu Kwaik. 1998. Identification of macrophage-specific infectivity loci (mil) of Legionella pneumophila that are not required for infectivity of protozoa. Infect. Immun. 66883-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao, L.-Y., O. S. Harb, and Y. Abu Kwaik. 1997. Utilization of similar mechanisms by Legionella pneumophila to parasitize two evolutionarily distant hosts, mammalian and protozoan cells. Infect. Immun. 654738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garduno, R. A., E. Garduno, M. Hiltz, and P. S. Hoffman. 2002. Intracellular growth of Legionella pneumophila gives rise to a differentiated form dissimilar to stationary-phase forms. Infect. Immun. 706273-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garduno, R. A., E. Garduno, and P. S. Hoffman. 1998. Surface-associated Hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 664602-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautier, L., L. Cope, B. M. Bolstad, and R. A. Irizarry. 2004. affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20307-315. [DOI] [PubMed] [Google Scholar]
- 31.Gunn, J. S. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16284-290. [DOI] [PubMed] [Google Scholar]
- 32.Habyarimana, F., S. Al-Khodor, A. Kalia, J. E. Graham, C. T. Price, M. T. Garcia, and Y. A. Kwaik. 2008. Role for the ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ. Microbiol. 101460-1474. [DOI] [PubMed] [Google Scholar]
- 33.Hammer, B. K., and M. S. Swanson. 1999. Co-ordination of legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol. Microbiol. 33721-731. [DOI] [PubMed] [Google Scholar]
- 34.Hammer, B. K., E. S. Tateda, and M. S. Swanson. 2002. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol. Microbiol. 44107-118. [DOI] [PubMed] [Google Scholar]
- 35.Harb, O. S., and Y. Abu Kwaik. 1999. Probing the microenvironment of intracellular bacterial pathogens. Microb. Infect. 1445-453. [DOI] [PubMed] [Google Scholar]
- 36.Harb, O. S., L. Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2251-265. [DOI] [PubMed] [Google Scholar]
- 37.Heeb, S., and D. Haas. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other gram-negative bacteria. Mol. Plant-Microbe Interact. 141351-1363. [DOI] [PubMed] [Google Scholar]
- 38.Hiltz, M. F., G. R. Sisson, A. K. Brassinga, E. Garduno, R. A. Garduno, and P. S. Hoffman. 2004. Expression of magA in Legionella pneumophila Philadelphia-1 is developmentally regulated and a marker of formation of mature intracellular forms. J. Bacteriol. 1863038-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3165-170. [DOI] [PubMed] [Google Scholar]
- 40.Holden, E. P., H. H. Winkler, D. O. Wood, and E. D. Leinbach. 1984. Intracellular growth of Legionella pneumophila within Acanthamoeba castellanii Neff. Infect. Immun. 4518-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 1581319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 1582108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Investig. 66441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jules, M., and C. Buchrieser. 2007. Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics. FEBS Lett. 5812829-2838. [DOI] [PubMed] [Google Scholar]
- 45.Kagan, J. C., and C. R. Roy. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4945-954. [DOI] [PubMed] [Google Scholar]
- 46.Kagan, J. C., M. P. Stein, M. Pypaert, and C. R. Roy. 2004. Legionella subvert the functions of rab1 and sec22b to create a replicative organelle. J. Exp. Med. 1991201-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lynch, D., N. Fieser, K. Gloggler, V. Forsbach-Birk, and R. Marre. 2003. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol. Lett. 219241-248. [DOI] [PubMed] [Google Scholar]
- 48.McPhee, J. B., M. Bains, G. Winsor, S. Lewenza, A. Kwasnicka, M. D. Brazas, F. S. Brinkman, and R. E. Hancock. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 1883995-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizuno, T. 1997. Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4161-168. [DOI] [PubMed] [Google Scholar]
- 50.Molmeret, M., D. Bitar, L. Han, and Y. Abu Kwaik. 2004. Disruption of the phagosomal membrane and egress of Legionella pneumophila into the cytoplasm during late stages of the intracellular infection of macrophages and Acanthamoeba polyphaga. Infect. Immun. 724040-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molmeret, M., M. Horn, M. Wagner, M. Santic, and Y. Abu Kwaik. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 7120-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Molofsky, A. B., and M. S. Swanson. 2003. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol. Microbiol. 50445-461. [DOI] [PubMed] [Google Scholar]
- 53.Newton, H. J., F. M. Sansom, J. Dao, C. Cazalet, H. Bruggemann, C. Albert-Weissenberger, C. Buchrieser, N. P. Cianciotto, and E. L. Hartland. 2008. Significant role for ladC in initiation of Legionella pneumophila infection. Infect. Immun. 763075-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 692569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez, J. C., and E. A. Groisman. 2007. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol. Microbiol. 63283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robey, M., and N. P. Cianciotto. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 705659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roland, K. L., L. E. Martin, C. R. Esther, and J. K. Spitznagel. 1993. Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with a possible role in virulence. J. Bacteriol. 1754154-4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowbotham, T. J. 1983. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roy, C. R., K. H. Berger, and R. R. Isberg. 1998. Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol. Microbiol. 28663-674. [DOI] [PubMed] [Google Scholar]
- 60.Santic, M., M. Molmeret, and Y. Abu Kwaik. 2005. Maturation of the Legionella pneumophila-containing phagosome into a phagolysosome within gamma interferon-activated macrophages. Infect. Immun. 733166-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sauer, J. D., M. A. Bachman, and M. S. Swanson. 2005. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. USA 1029924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Segal, G., M. Feldman, and T. Zusman. 2005. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol. Rev. 2965-81. [DOI] [PubMed] [Google Scholar]
- 63.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 672117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3178-185. [DOI] [PubMed] [Google Scholar]
- 65.Steinert, M., K. Heuner, C. Buchrieser, C. Albert-Weissenberger, and G. Glockner. 2007. Legionella pathogenicity: genome structure, regulatory networks, and the host cell response. Int. J. Med. Microbiol. 297577-587. [DOI] [PubMed] [Google Scholar]
- 66.Stone, B. J., and Y. Abu Kwaik. 1998. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells Infect. Immun. 661768-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competency for DNA uptake by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 1811395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stone, B. J., A. Brier, and Y. A. Kwaik. 1999. The Legionella pneumophila prp locus; required during infection of macrophages and amoebae. Microb. Pathog. 27369-376. [DOI] [PubMed] [Google Scholar]
- 69.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 1009440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 633609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tilney, L. G., O. S. Harb, P. S. Connelly, C. G. Robinson, and C. R. Roy. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 1144637-4650. [DOI] [PubMed] [Google Scholar]
- 72.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tyndall, R. L., and E. L. Domingue. 1982. Cocultivation of Legionella pneumophila and free-living amoebae. Appl. Environ. Microbiol. 44954-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279873-876. [DOI] [PubMed] [Google Scholar]
- 75.Vogel, J. P., and R. R. Isberg. 1999. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 230-34. [DOI] [PubMed] [Google Scholar]
- 76.Weeratna, R., T. J. Marrie, S. M. Logan, D. Hoskin, P. S. Hoffman, L. Yates, S. Burbridge, D. Haldane, and G. Bezanson. 1993. Legionnaires' disease in cardiac transplant patients: a cell-mediated immune response develops despite cyclosporine therapy. J. Infect. Dis. 168521-522. [DOI] [PubMed] [Google Scholar]
- 77.Winn, W. C., Jr. 1988. Legionnaires disease: historical perspective. Clin. Microbiol. Rev. 160-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wosten, M. M., L. F. Kox, S. Chamnongpol, F. C. Soncini, and E. A. Groisman. 2000. A signal transduction system that responds to extracellular iron. Cell 103113-125. [DOI] [PubMed] [Google Scholar]
- 79.Zusman, T., G. Aloni, E. Halperin, H. Kotzer, E. Degtyar, M. Feldman, and G. Segal. 2007. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol. Microbiol. 631508-1523. [DOI] [PubMed] [Google Scholar]
- 80.Zusman, T., O. Gal-Mor, and G. Segal. 2002. Characterization of a Legionella pneumophila relA insertion mutant and roles of RelA and RpoS in virulence gene expression. J. Bacteriol. 18467-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.