Abstract
Infection of humans with Ehrlichia chaffeensis, the etiologic agent of human monocytic ehrlichiosis, can cause hepatitis of various levels of severity. When the three human isolates of E. chaffeensis, each belonging to a different genogroup, are inoculated into severe combined immunodeficiency mice, the order of severity of clinical signs and bacterial burden detected in the liver is as follows (from greatest to least severity and highest to lowest burden): strain Wakulla, followed by strain Liberty, followed by strain Arkansas. In this article, we used microarray analysis to define transcriptional profiles characteristic of the histopathological features in the mouse liver. Cytokine and chemokine profiles and their receptor profiles were strikingly different among the three strains of E. chaffeensis: gamma interferon, CCL5, CXCL1, CXCL2, CXCL7, CXCL9, interleukin 2 receptor gamma (IL2Rγ), IL21R, CCR2, and CXCR6 were highly upregulated with strain Arkansas; and tumor necrosis factor (TNF), CCL2, CCL3, CCL5, CCL6, CCL12, CCL20, CXCL2, CXCL7, CXCL9, CXCL13, TNF receptor superfamily 9 (TNFRSF9), TNFRSF13β, IL1R2, IL2Rγ, IL20Rβ, IL21R, CCR1, CCR2, and CXCR4 were highly upregulated with strain Wakulla. With strain Liberty, only CXCL13 was highly upregulated, and IL13Rα2 was downregulated. In livers infected with the Arkansas strain, monocytes/macrophages and NK cells were enriched in the granulomas and an increase in NK cell marker mRNAs was detected. Livers infected with the Wakulla strain displayed infiltration of significantly more neutrophils and an increase in neutrophil marker mRNAs. Genes commonly upregulated in liver tissue infected with the three strains are other host innate immune and inflammatory response genes, including those encoding several acute-phase proteins. Genes downregulated commonly are related to host physiologic functions. The results suggest that marked modulation of host cytokine and chemokine profiles by E. chaffeensis strains underlies the distinct host liver disease.
Human monocytic ehrlichiosis (HME) was discovered in 1986 (34), and the etiologic agent, a monocyte-tropic Ehrlichia species, was isolated several years later and named Ehrlichia chaffeensis (15). HME is one of the most prevalent life-threatening tick-borne zoonoses in North America and was designated a nationally notifiable disease in 1998 (35). HME is an acute febrile illness characterized by headache, malaise, nausea, and myalgia and/or arthralgia and frequently accompanied by leukopenia, thrombocytopenia, anemia, and elevation of hepatic transaminase levels (43). The severity of HME, however, varies from subclinical infection to death. A previous investigation of the pathology of injury in liver tissues from five immunocompetent patients, one human immunodeficiency virus (HIV)-infected patient, and one monoclonal gammopathy patient with laboratory-confirmed HME revealed scattered lobular lymphohistiocytic foci and diffuse lymphohistiocytic infiltration and Kupffer cell hyperplasia with moderate cholestasis (cholestasis was not noted in the HIV patient) (56). The inflammation was not correlated with the levels of infection (56). Nonetheless, little is known about pathogenesis of HME liver injury. Although E. chaffeensis is a gram-negative bacterium, belonging to the order Rickettsiales, well-characterized bacterial virulence factors, such as exotoxins, endotoxins, capsule polysaccharides or proteins, adherence pili, or virulence plasmids, have not been detected in E. chaffeensis. Conventional approaches to identifying virulence determinants that involve transformations and mutations are not readily available for obligatory intracellular bacteria. Another classical way to identify bacterial virulence determinants is to compare virulent and avirulent strains. The genes that vary between the two strains are more likely to encode virulence factors than the genes that are highly conserved between them. Our previous study of comparative genome hybridization showed genes varying among three E. chaffeensis strains of diverse virulence (36). Importantly, most of these genes encode putative membrane proteins and hypothetical proteins, suggesting a novel pathogenic mechanism. Dissection of the relative roles of the pathogen strain versus host factors is very difficult with human infections due to the relatively low number of clinical cases and laboratory tissue specimens with a full data set and unknowns regarding the strain type in most infections. The use of these three previously characterized E. chaffeensis strains (36) in a single experimental host species allows testing of whether there are intrinsic differences in the response to each strain.
The immune system of vertebrates is composed of two major components: innate immunity and adaptive immunity (25). Patients with HME may develop a fulminant toxic or septic shock-like syndrome, particularly individuals with HIV infection or other immune deficiencies (e.g., organ transplant recipients or patients undergoing immunosuppressive treatment for cancer therapy or for immune disorders) (43). Although immunocompetent mice clear E. chaffeensis Arkansas infection within 2 weeks and do not develop clinical signs, infection of severe combined immunodeficiency (SCID) mice with the Arkansas strain induces fatal illness accompanied by severe granulomatous hepatic inflammation similar to those seen in some HME patients (63). These studies suggest the role of the innate immune response in HME pathogenesis.
We previously selected the Wakulla (group II) and Liberty (group III) strains of E. chaffeensis and compared their pathogenesis and genomic sequences with those of the Arkansas strain (group I) (group designation is based on the work of Cheng et al. [10]) (36). To compare pathogenesis, we inoculated these isolates into SCID mice. The numbers of Wakulla and Liberty bacteria in the blood increased about 90- and 60-fold, respectively, from day 5 to 15 postinfection (p.i.), whereas Arkansas increased only 3-fold. The livers of Wakulla- and Liberty-inoculated mice had approximately 900- and 10-fold more bacteria, respectively, than the livers of mice inoculated with Arkansas. Granulomatous lymphohistiocytic infiltration in the liver was prominent with Arkansas infection, and Wakulla infection induced profound and diffuse lymphohistiocytic infiltration. Atrophy and focal necrosis of parenchymal cells were evident in the livers of mice inoculated with Wakulla. The livers of mice inoculated with Liberty were almost free from these hepatic lesions. Serum aspartate transaminase activity at day 15 p.i. was significantly elevated in the sera of mice infected with Wakulla (36). These results indicate that the diverse virulence and pathogenesis patterns of E. chaffeensis strains Wakulla and Liberty were distinct from each other and from those of Arkansas. Because the range of liver pathology in the SCID mice mimics that seen in HME patients (43), the SCID mouse model can be used to study a molecular signature for host innate responses to E. chaffeensis and to illustrate both common and distinct global virulence effects of highly virulent and less-virulent strains.
Differences in transcriptome profiles of infected and uninfected human monocytes are indicative of virulence determinant functions. Transcriptomes of human monocytes infected with E. chaffeensis have not been studied. However, E. chaffeensis Arkansas infection in a human acute leukemia monocyte cell line (THP-1) significantly alters the transcriptional levels of 4.5% of host genes, including those encoding apoptosis inhibitors, proteins regulating cell differentiation, signal transduction, proinflammatory cytokines, biosynthetic and metabolic proteins, and membrane trafficking proteins at 1 to 24 h postinoculation (65). Transcriptome analysis also may elucidate mechanisms that lead to difference in disease status. For example, using whole blood incubated with meningococci in vitro, gene expression profiling combined with protein and cellular methods identified interleukin 6 as the cause for significant myocardial depression (44). Gene transcriptional changes in the mouse lung infected with Francisella tularensis and stomachs from mice infected with Helicobacter species uncovered molecular markers of each disease (1, 38, 39). In the present study, we examined global gene expression profiles in the livers of the SCID mice infected with the three E. chaffeensis strains at day 15 p.i. Here we present data indicating that the common and distinct innate immune and inflammatory responses to highly virulent and less-virulent strains occur locally in the liver environment and are clearly reflected at the gene expression level.
MATERIALS AND METHODS
Bacteria and mice.
The Arkansas, Wakulla, and Liberty strains of E. chaffeensis were propagated in DH82 cells as described previously (36). Three- to four-week-old male ICR-SCID mice were obtained from Taconic (Tarrytown, NY) and injected intraperitoneally with 106 E. chaffeensis-infected DH82 cells (typically >95% infected) containing approximately the same number of bacteria or 106 uninfected DH82 cells (mock). The number of bacteria was estimated as described previously (64). The correlation coefficient (r) value between the bacterial genome equivalent based on real-time PCR and the bacterial count under a light microscope was 0.932 (36). At 15 days p.i., mice were euthanized and livers were harvested and fixed in Histochoice (Amresco, Solon, OH) for histopathological examination or were stored in RNAlater (Qiagen, Valencia, CA) for microarray and real-time reverse transcription (RT)-PCR analyses. Animal experimentation protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Ohio State University.
Microarray analysis.
Total RNA was extracted from E. chaffeensis-infected or uninfected mouse livers using the RNeasy mini-kit (Qiagen). Double-stranded cDNA was prepared from approximately 8 μg total RNA using the GeneChip One-Cycle cDNA synthesis kit (Affymetrix, Santa Clara, CA). Biotin-labeled cRNA was generated from the cDNA using the GeneChip IVT labeling kit (Affymetrix). The labeled cRNA was hybridized to a GeneChip Mouse Genome 430 2.0 array (Affymetrix) spotted with approximately 39,000 murine cDNA elements. After washing and staining with streptavidin phycoerythrin were carried out, the array was scanned using a GeneChip 3000 7G scanner (Affymetrix). Data analysis was performed using the Partek Discovery Suite software program (Partek Inc., St. Louis, MO). The average fluorescence intensity of each probe obtained from triplicate experiments derived from three liver samples per strain of E. chaffeensis was compared with the average fluorescence intensity obtained using triplicate sham-infected liver samples to generate the difference in intensity for each gene call. For each probe, when the difference between the average fluorescence intensity from three E. chaffeensis-infected mice and that from three sham-infected mice gave a P value of <0.05 by analysis of variance (ANOVA), it was considered significant. In addition, for each probe, when the log2 value of the ratio of the average fluorescence intensity from three E. chaffeensis-infected mice to that from three sham-infected mice was a >2-fold or <2-fold standard deviation of the mean for all detected probes, it was considered upregulated or downregulated, respectively. Standard deviations of data of strains Liberty, Arkansas, and Wakulla were 1.54, 1.61, and 1.70, respectively. Probes were annotated using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/) or the NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx).
Real-time RT-PCR.
Total RNA was extracted from liver specimens using the RNeasy kit (Qiagen). cDNAs were prepared from 1 μg total RNA using SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) and an oligo(dT) primer. The cDNA concentrations of selected genes were quantified by real-time PCR using gene-specific primers (see Table S1 in the supplemental material).
Immunohistochemistry.
Sequential 4-μm paraffin sections were stained with an antibody specific for Kupffer cells and macrophages (F4/80; AbD Serotec, Raleigh, NC) or with an antibody specific for natural killer (NK) cells (asialo GM1; Cedarlane, Hornby, Canada). Detection was performed using biotinylated secondary antibodies in combination with horseradish peroxidase-coupled streptavidin (Jackson ImmunoResearch, West Grove, PA) and the substrate DAB (Research Genetics/Invitrogen). Sections were also stained with Giemsa (Dako, Carpinteria, CA). As negative controls, normal rabbit serum and isotype-matched mouse antibody (immunoglobulin G2b) were used. All sections were counterstained with hematoxylin. Photographic images were obtained using a Nikon Eclipse 50i microscope (Nikon Instruments Inc., Melville, NY).
Statistical analysis.
One-way ANOVA with the Tukey honestly significant difference test was performed to determine the significance of differences between groups. A P value of <0.05 was considered significant.
Microarray data accession number.
The NCBI Gene Expression Omnibus accession no. of the microarray data is GSE8966.
RESULTS
Immunohistochemical analysis.
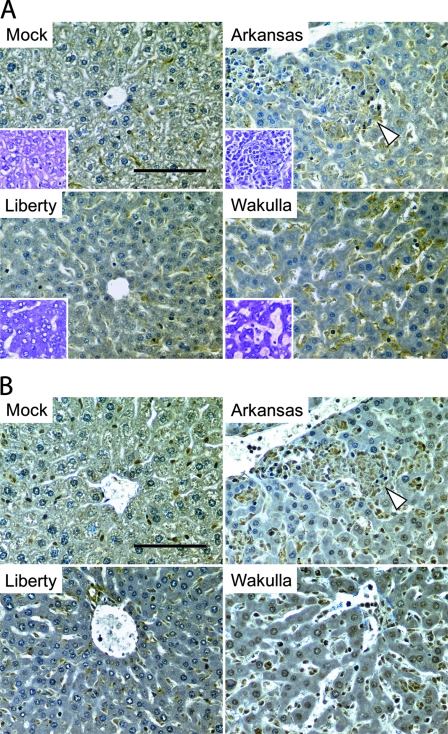
In order to examine the types of infiltrated leukocytes in liver tissues of E. chaffeensis-infected SCID mice, at day 15 p.i. sequential 4-μm sections of paraffin-embedded liver tissues were immunoperoxidase stained using anti-F4/80 antibody for monocytes/macrophages or anti-asialo GM1 antibody for NK cells (Fig. 1). Giemsa staining was used to differentiate neutrophils (polymorphic nuclei) from other types of cells. While overall an increase in monocytes/macrophages was detected in liver tissues of mice infected with all three strains of E. chaffeensis, there was a significant increase in monocytes/macrophages in the granulomas of strain Arkansas-infected livers (Fig. 1A and D). Many cells in the sinusoid lumen in Wakulla-infected livers and the granulomas of Arkansas-infected livers were stained strongly with anti-asialo GM1 antibody (Fig. 1B and D). Fewer cells in the sinusoid lumen of Liberty-infected livers stained positive with anti-asialo GM1, but the staining was still significant relative to results for the control uninfected group. A significant amount of neutrophil infiltration was detected in Wakulla-infected livers (Fig. 1C and D). Neutrophil infiltration was less prevalent in the granuloma of Arkansas-infected livers, and very few neutrophils were found in Liberty-infected livers. In contrast, neutrophils were rarely found in mock-inoculated liver tissues.
FIG. 1.
Immunohistochemistry of livers infected with three strains of E. chaffeensis. (A) Kupffer cells and macrophages/monocytes in liver specimens from mock- or E. chaffeensis-infected mice immunolabeled with anti-F4/80 antibody. Inserts are hematoxylin-stained livers. (B) NK cells in liver specimens from mock- or E. chaffeensis-infected mice immunolabeled with anti-asialo GM1 antibody. (C) Leukocyte infiltration in liver specimens from mock- or E. chaffeensis-infected mice using Giemsa staining. Insert shows neutrophils at higher magnification (×1,000). (A to C) White arrowheads indicate granulomas. Bar = 100 μm. (D) Relative numbers of macrophages/monocytes, NK cells, and neutrophils per unit area in liver tissues from mock- or E. chaffeensis-infected mice. Cell numbers were determined by counting immuno- or Giemsa-stained cells in five randomly chosen fields (0.79 mm2) each from Liberty (Lib), Arkansas (Ark), and Wakulla (Wak)-infected livers without granuloma (D) or within granulomas (G), and the mean values were normalized to the mean cell numbers from mock (Moc)-infected mice. **, P < 0.01 (ANOVA).
Gene expression profiling in hepatic tissues.
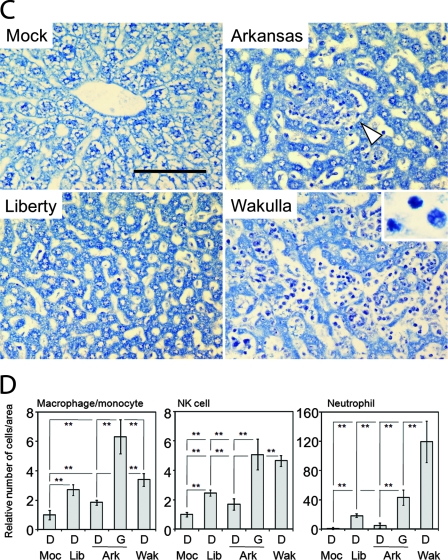
Global gene expression profiles were determined for liver specimens from the following four groups of three mice each at day 15 p.i.: a group infected with E. chaffeensis Arkansas, a group infected with E. chaffeensis Liberty, a group infected with E. chaffeensis Wakulla, and a control (mock) group inoculated with uninfected DH82 cells (host cells used to cultivate E. chaffeensis). Using the stringent cutoff criterion described in Materials and Methods, which corresponds to an approximately eightfold signal intensity difference from the control, a total of 776 mouse genes were highly up- or downregulated. The number of upregulated mouse genes was greatest for Wakulla, followed by Arkansas, followed by Liberty, and most up- or downregulated mouse genes were specific to each bacterial strain (Fig. 2). Wakulla and Arkansas shared more upregulated mouse genes than Liberty and Arkansas or Liberty and Wakulla (Fig. 2). Wakulla and Liberty had more downregulated mouse genes than Arkansas had (Fig. 2). Details of the 776 genes categorized in Fig. 2, including signal intensity ratios and statistical analysis values, are shown in Table S2 in the supplemental material.
FIG. 2.
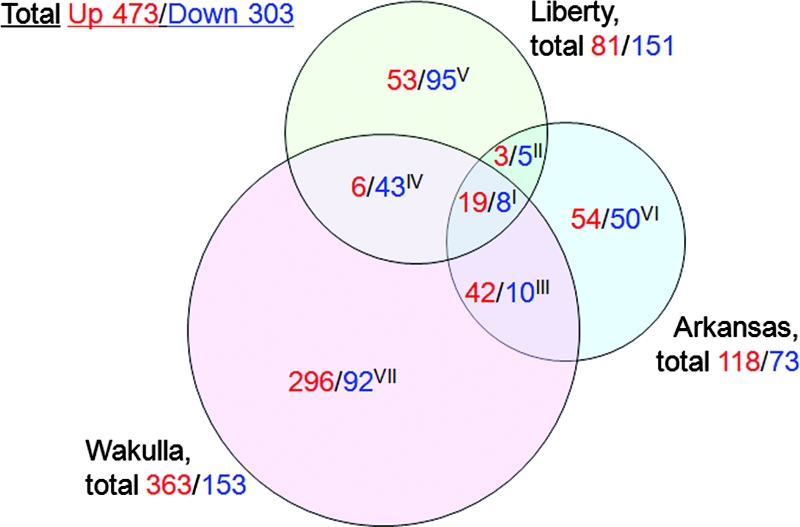
Comparison of liver transcriptome from mice infected with three strains of E. chaffeensis. The numbers of mouse genes upregulated (red) or downregulated (green) with three strains of E. chaffeensis are shown. Numbers within intersection of circles indicate genes shared by two or three strains. Genes belonging to sections I to VII are shown in Table S2 in the supplemental material.
Inflammatory and immune gene expression.
Microarray data analysis using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/) showed that 76, 35, and 13 genes up- or downregulated by infection with Wakulla, Arkansas, and Liberty, respectively, belong to the immune and/or inflammatory response functional category (Fig. 3A) (see Table S3 in the supplemental material). Among immune and/or inflammatory response functional category genes, cytokine and chemokine (descriptions are mostly based on the work in reference 9) profiles were strikingly different among three strains of E. chaffeensis. Gamma interferon (IFN-γ) (which is secreted by Th1 cells, cytotoxic T cells, dendritic cells, and NK cells and activates monocytes/macrophages) was significantly upregulated in mice infected with the Arkansas strain (Fig. 3B). This suggests that the Arkansas strain induced IFN-γ production by NK or dendritic cells in the infected mouse liver. The gene encoding tumor necrosis factor (TNF) (which is mainly produced by macrophages, regulates immune cells, and is involves in systemic inflammation) was highly upregulated with strain Wakulla (Fig. 3B). In contrast, IFN-γ and TNF were not highly upregulated with strain Liberty.
FIG. 3.
Expression of cytokine and chemokine genes in E. chaffeensis-infected livers. (A) Numbers of mouse genes belonging to immune and/or inflammatory response category (by functional annotation tool of DAVID Bioinformatics Resources) differentially expressed by E. chaffeensis infection. (B to E) The heat maps show relative expression in log2 values (infected/uninfected). A tile with a diagonal line shows two distinct probes for the same gene. (B) Differentially expressed cytokine and chemokine genes and their receptor genes. Asterisks indicate genes confirmed by RT-PCR (see Fig. 4). (C) Differentially expressed IFN-inducible genes. (D) Differentially expressed genes encoding cell markers. (E) Commonly upregulated (red) and downregulated (green) genes in mice infected with three E. chaffeensis strains (see section I in Fig. 2). Genes with the highest average ratios are shown from the top. Asterisks indicate acute-phase proteins. Gray tile in panels B, C, and D indicates the ratio under the threshold or no significant difference between infected and uninfected livers.
Chemokines are a large family of specialized heparin-binding proteins, the primary and traditional function of which is to regulate the trafficking of leukocytes. The chemokine system is comprised of ∼50 molecules and 20 receptors in humans, with orthologs in other mammalian species (52). The chemokine ligand superfamily is divided into subgroups, of which the largest are the CC (C-C motif) chemokines (28 members) and the CXC (C-X-C motif) chemokines (16 members). Chemokine subgroup members, encoded in multigene arrays, are functionally related and signal to corresponding families of chemokine receptors (37). A large number of chemokine genes attracting monocytes and neutrophils were strongly upregulated with Wakulla, followed by the Arkansas strain. With strain Wakulla, CCL2 (which recruits monocytes, memory T cells, and dendritic cells), CCL3 (which is mainly produced by macrophages and activates granulocytes), CCL5 (which is chemotactic to T cells and monocytes), CCL6 (which is expressed by myeloid cells and attracts monocytes), CCL12 (which is expressed by macrophages and attracts eosinophils, monocytes, and lymphocytes), CCL20, also called liver activation-regulated chemokine (which chemoattracts lymphocytes and dendritic cells), CXCL2 (which is secreted by monocytes and macrophages and is chemotactic for polymorphonuclear leukocytes and hematopoietic stem cells), CXCL7, also called Ppbp (proplatelet basic protein) (released in large amounts from platelets following their activation), CXCL9 (a T-cell chemoattractant, induced by IFN-γ), and CXCL13 (which is secreted by dendritic cells and attracts B cells) were upregulated (Fig. 3B). With the Arkansas strain CCL5, CXCL1 (which is expressed by macrophages, neutrophils, and epithelial cells and has neutrophil chemoattractant activity), CXCL2, CXCL7, and CXCL9 were significantly upregulated (Fig. 3B). Among these genes, CCL5, CXCL2, CXCL7, and CXCL9 were commonly upregulated in mice infected with the Arkansas and Wakulla strains (Fig. 3B). By the stringent cutoff criterion defined in this study, chemokine genes were not upregulated in liver tissues from Liberty-infected mice, except for the CXCL13 gene (Fig. 3B).
Among receptors of cytokines and chemokines, TNF receptor superfamily 9 (TNFRSF9) (CD137) (can be expressed by dendritic cells, NK cells, granulocytes, endothelial cells, and activated T cells at sites of inflammation) (18), TNFRSF13β (expressed by lymphocytes and monocytes/macrophages), interleukin 1 receptor, type II (IL1R2), also known as CD121b (expressed by monocytes and binds interleukin 1α [IL-1α]) and IL-1β and acts as a decoy receptor that inhibits the activity of its ligands) [12]), IL-2 receptor gamma (IL2Rγ) (involved in making the common gamma chain), IL-20 receptor β (IL20Rβ) (not expressed by cells of the hematopoietic lineage but expressed in various other tissues [40]), IL-21 receptor (IL21R) (expressed by NK-, T-, and B-cell lines [42]), CCR1 (expressed by T cells, monocytes, eosinophils, and basophils [9], and ligands of this receptor include CCL3, CCL5, CCL7 [monocyte chemoattractant protein 3], and CCL24 [myeloid progenitor inhibitory factor 1]), CCR2 (expressed by monocytes, dendritic cells, and memory T cells, and the receptor for CCL2, CCL8, CCL7, CCL13, and CCL16 [9]), and CXCR4 (CXCL12 receptor) were highly upregulated with strain Wakulla (Fig. 3B). IL2Rγ, IL21R, CCR2, and CXCR6 (receptor for CXCL16, expressed by NK cells [9]) were highly upregulated in livers of mice infected with strain Arkansas. In livers of mice infected with strain Liberty, no genes encoding receptors of cytokines and chemokines were highly upregulated, while IL-13 receptor alpha 2 (expressed by macrophages [19]) was downregulated (Fig. 3B). IL2Rγ and CXCR4 genes are widely expressed in several types of cells.
In order to include moderately up- or downregulated cytokine and chemokine genes in the analysis, genes below our cutoff criterion (an approximately eightfold signal difference compared with results for the control liver) but with a significant difference (P < 0.05) were examined (see Table S4 in the supplemental material). Among cytokines and chemokines in this category, in liver tissues from Arkansas-infected mice, expression of CXCL10 (produced by monocytes, endothelial cells, and fibroblasts in response to IFN-γ and attracts monocytes/macrophages, NK cells, dendritic cells, and T cells) and CXCL11 (induced by IFN-γ and beta interferon and attracts T cells) was significantly greater than in that liver tissues from uninfected mice or from mice infected with the other two strains. The immunosuppressive cytokines IL-10 (produced primarily by monocytes) and transforming growth factor β1 (TGF-β1) (48), TGF-β3 (regulates molecules involved in cellular adhesion and extracellular matrix), and IL-1β were significantly upregulated only in Wakulla-infected livers (see Table S5 in the supplemental material).
Among the IFN-inducible genes, three genes upregulated only with the Wakulla strain, the Ifit2 (interferon-induced protein with tetratricopeptide repeats 2) (5), Mx1 (myxovirus [influenza virus] resistance 1) (4), and Ifi44 (interferon-induced protein 44) (27) genes, are alpha interferon-regulated genes (Fig. 3D). In contrast, IFN-γ-induced histocompatibility 2, class II antigen E beta, and class II transactivator were upregulated only in mice infected with the Arkansas strain (Fig. 3C).
Inflammatory cell type-specific genes.
The NK cell markers CD7 (47) and Klrb1f (killer cell lectin-like receptor subfamily B member 1F) (45) were upregulated in livers from Arkansas-infected mice (Fig. 3D), although the number of infiltrating NK cells in granulomas in livers from Arkansas-infected mice was similar to that in livers from Wakulla-infected mice (Fig. 1B and D). Macrophage and Kupffer cell markers (CD14 and G6pd2/G6pdx) (50, 58) were upregulated only in Wakulla-infected livers (Fig. 3D). Neutrophil markers (Ela2, Itgb2, Ltf, and Fcgr1) (16, 31, 41, 54) were upregulated only in Wakulla-infected livers (Fig. 3D), in agreement with a large number of infiltrating neutrophils in the liver tissues (Fig. 1C and D).
Commonly up- and downregulated genes.
Of the 19 upregulated genes common to mice infected with the three E. chaffeensis strains (Fig. 2 and 3E), eight genes, encoding Orm3, Orm2, Serpina3G, Gzma, Gbp2, Saa3, Tgtp, and H2-DMb2, belong to the immune and/or inflammatory response functional category (Fig. 3A). The Orm2 (orosomucoid 2), Orm3 (orosomucoid 3), Lcn2 (lipocalin 2), and Saa3 (serum amyloid A3) genes encode acute-phase proteins (23, 30, 59). In addition, other genes relating to innate immune and inflammatory responses, including those encoding macrophage receptor with collagenous structure (Marco), metallothioneins (Mt1 and Mt2), serine peptidase inhibitor, clade A, member 3G (Serpina3g) (21), fibrinogen-like protein 2 (Fgl2), and lysozyme, were commonly upregulated (Fig. 3E).
Eight genes, including those encoding Slc22a7 (solute carrier family 22, member 7), preferentially expressed in the liver (57), Ddc (dopa decarboxylase), Elovl3 (an enzyme involved in elongation of very long chain fatty acids), and Hsd3β5 (3-beta-hydroxy-delta5-steroid dehydrogenase 5, a key enzyme in steroid hormone metabolism) were commonly downregulated in liver tissues with the three strains of E. chaffeensis (Fig. 3E). Downregulation of these genes suggests a physiological change in hepatic parenchymal cells.
Real time RT-PCR analysis.
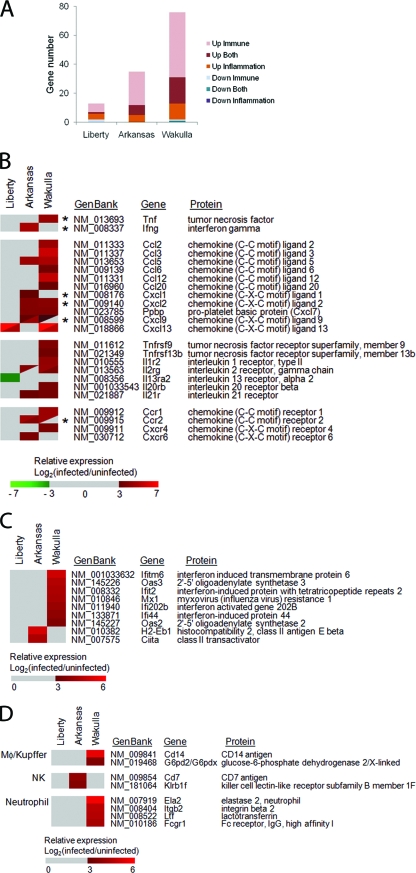
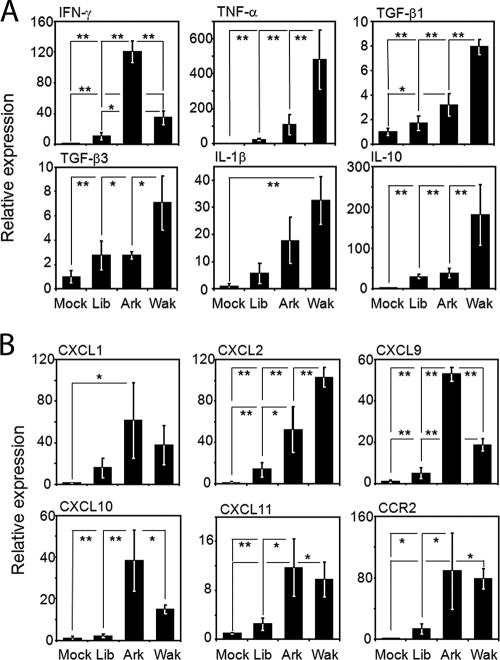
Differential expression of six genes shown in Fig. 3B and six genes shown in Table S5 in the supplemental material among mice infected with each of three E. chaffeensis strains in the microarray analysis were confirmed using real-time RT-PCR (Fig. 4). Most of the results were consistent with those of the cDNA microarray analysis. In liver tissues from Arkansas-infected mice, expression of IFN-γ, CXCL9, and CXCL10 was significantly higher than that in liver tissues from mice infected with the other two strains. Similarly, in liver tissue from Wakulla-infected mice, expression of IL-10, CXCL2, TNF, and TGF-β was significantly higher. Expression of CXCL11 and CCR2 was significantly higher in Arkansas- or Wakulla-infected livers than in mock-infected livers (Fig. 4). Expression of IL-1β was significantly high only in Wakulla-infected livers in comparison with results for uninfected livers. In liver tissue from Liberty-infected mice, expression of every cytokine and chemokine gene determined by the real-time RT-PCR was significantly lower than that in mice infected with the other strains, although most of the cytokine and chemokine gene expression levels were slightly higher than those from uninfected livers (Fig. 4).
FIG. 4.
Real-time RT-PCR analysis of differentially expressed cytokine- and chemokine-related genes in E. chaffeensis-infected livers. (A) Cytokine genes. (B) Chemokine genes. The mean expression level of each gene in liver tissues from E. chaffeensis-infected mice relative to the mean expression in liver tissues from mock-infected mice is shown. All data are normalized to glyceraldehyde-3-dehydrogenase expression. **, P < 0.01; *, P < 0.05 (ANOVA). Ark, Arkansas strain; Lib, Liberty strain; Wak, Wakulla strain.
DISCUSSION
The SCID mouse lacks mature T and B cells but has remaining cells involved in immunity and inflammation, including monocytes/macrophages, neutrophils, NK cells, and dendritic cells, which are capable of producing cytokines and chemokines in response to pathogens (6, 14). The SCID mouse is the only animal model with which E. chaffeensis strain-dependent pathogenesis has been reported (36). Since an immunocompetent mouse model is currently unavailable, the SCID mouse is the best in vivo model for comparing host immune and inflammatory responses to different E. chaffeensis strains. IFN-γ, produced by activated T cells and NK cells in immunocompetent hosts, is one of the most important known factors for the host in combating E. chaffeensis infection. Exogenous IFN-γ treatment of human peripheral blood monocytes significantly inhibits E. chaffeensis Arkansas infection in vitro (3). IFN-γ has been shown to provide a protective role in immunocompetent mice infected with the Ehrlichia HF strain (an Ixodes ovatus-isolated Ehrlichia strain) (24). The present data showed that in livers from SCID mice infected with the Arkansas strain, IFN-γ was highly upregulated. In SCID mice, due to a lack of mature T cells, the source of IFN-γ is mainly activated NK cells. Consistent with this observation, NK cell marker genes were upregulated in the livers from strain Arkansas-infected mice. The Arkansas strain produces a smaller bacterial load and less-severe clinical signs in mice than the Liberty or Wakulla strain (36). Therefore, the present data suggest that strong induction of IFN-γ in NK cells upon infection with the Arkansas strain likely mediates the observed suppression of bacterial proliferation. In contrast, poor induction of IFN-γ in NK cells in SCID mice infected with the Liberty or Wakulla strain likely results in greater bacterial proliferation. IL-10 suppresses secretion of IFN-γ by NK cells (51), and TGF-β suppresses NK cell activation (62). Since IL-10 and TGF-β were significantly upregulated in Wakulla-infected liver tissues, this may explain the observed lack of IFN-γ upregulation in Wakulla-infected livers, even though infiltration of NK cells was observed. In contrast, expression of IFN-γ, IL-10, and TGF-β1 was remarkably lower in livers from Liberty-infected mice than in livers from mice infected with the other strains, in agreement with less change in IFN-regulated genes and in most of the immune and inflammatory gene expression with infection with the Liberty strain.
The three strains of E. chaffeensis induce strikingly different histopathologic lesions in liver tissue (36). Prominent granuloma development in the livers of Arkansas-infected mice is consistent with the upregulation of IFN-γ, because formation and resolution of granulomas involve IFN-γ (60). Neutrophil infiltration was observed in liver tissue from Arkansas- and Wakulla-infected mice but not in liver tissues from Liberty-infected mice. This is consistent with the upregulation of CXCL1, CXCL2, and CCL3, which mediates neutrophil recruitment (7) in liver tissues from Arkansas- and Wakulla-infected mice but not in those from Liberty-infected mice. Monocytes respond to CCL2 (32). While CCL2 receptor CCR2 was upregulated in liver tissues from Arkansas- and Wakulla-infected mice, CCL2 was upregulated in the liver tissues infected only with the Wakulla strain. Thus, the substantial infiltration of monocytes and neutrophils observed in liver tissues from Wakulla-infected mice and the minimal infiltration of leukocytes in livers from Liberty-infected mice may be attributed, in part, to the regulation by these chemokines and some of their receptors. CXCL9, CXCL10, and CXCL11 are induced by IFN-γ (13, 26, 33). These three chemokine genes were highly upregulated in Arkansas-infected livers, whereas none of them was significantly upregulated in Liberty-infected mice. Overall, low levels of most of the chemokines likely reduced infiltration of NK cells, macrophages, and neutrophils in livers infected with the Liberty strain.
Severe inflammatory cell infiltration, including neutrophils, in the livers of Wakulla-infected mice is also likely due to upregulation of TNF, TNFRSF9, and TNFRSF13β. TNFRSF9 and TNFRSF13β are expressed by monocytes (8, 55). TNF activates endothelial cells to express adhesion molecules and synthesize IL-1, IL-5, IL-6, IL-8, IL-11, monocyte chemoattractant protein 1, RANTES, granulocyte-macrophage colony-stimulating factor (CSF), granulocyte CSF, macrophage CSF (29), and lipid chemoattractants that are presented on their luminal surface (28, 46). Activated endothelial cells also transport chemoattractants from their luminal surface. Other chemoattractants can be generated by proteolytic cleavage in activated mast cells and platelets and delivered to endothelial cells through circulating microparticles or exocytosis of intracellular granules. For example, platelets are known to deposit CCL5, CXCL4, and CXCL7 onto the inflamed endothelium and thereby trigger the arrest of rolling monocytes to accelerate diapedisis (2, 61). Although the source of TNF is different from that in the present study (SCID mice), uncontrolled TNF production by CD8+ T cells was reported previously in association with immunopathology and failure to clear the Ixodes ovatus-isolated Ehrlichia strain in immunocompetent mice (24).
IL-1 and IL1R2 were strongly activated only with strain Wakulla. IL-1 acts on myelomonocytic cells through the type I receptor, and IL1R2, the type II receptor, inhibits IL-1 activity by acting as a decoy target for IL-1 (11). Thus, although IL-1 was induced, upregulation of the decoy receptor might have blocked the myeloid cell activation with strain Wakulla. IL2Rγ and IL21R were commonly upregulated with the Wakulla and Arkansas strains. IL20R1 and IL20R2 are absent or expressed at extremely low levels on cells of the hematopoietic lineage. Instead, these receptors are mainly found on epithelial and stromal cells and fibroblasts of various tissues (40). IL21R is important for the proliferation and differentiation of T cells, B cells, and NK cells. IL-21/IL21R plays a role in modulating innate and acquired effector mechanisms of murine macrophage by linking these different functions to support CD4+ T-cell-mediated immune responses (53). These changes in interleukin receptor expression and cytokines that activate or suppress T or B cells, combined with changes in expression of chemokines that attract T or B cells, are expected to influence the subsequent adaptive immune responses. Thus, future study using an immunocompetent animal model may yield more-complete insights into immune and inflammatory responses in immunocompetent and immunocompromised HME patients.
We previously reported the detection of several acute-phase proteins in the blood of dogs experimentally infected with Ehrlichia canis (49). Acute-phase proteins are a class of proteins whose plasma concentrations increase or decrease in response to inflammation. Some act to destroy or inhibit growth of microbes; others give negative feedback on the inflammatory response or are involved in pathogenesis (17, 22). Several acute-phase proteins were highly upregulated by infection with all three E. chaffeensis strains. Among acute-phase proteins, lipocalin 2, an antimicrobial peptide secreted by neutrophils or macrophages, captures iron-laden microbial siderophores (20). Whether any of these acute-phase proteins and other host innate immune response proteins commonly upregulated with three strains of E. chaffeensis have antiehrlichial activity in murine and/or human hosts awaits further investigation.
Taken together, the transcriptional profiling revealed specific gene expression patterns associated with distinct histopathologic features of liver specimens from mice infected with highly virulent and less-virulent strains of E. chaffeensis. The results indicate that pathogenicity of E. chaffeensis resides in an inability to suppress an excessive or inappropriate innate immune response. This is the first report on the infected tissue transcriptome profile in the order Rickettsiales. Since the transcriptional responses to E. chaffeensis could be dynamic and dose dependent, future in vitro comparative studies of host cell responses to different strains of E. chaffeensis or to their potential virulence factors, including dosages and temporal responses, would provide more insights into E. chaffeensis intrinsic factors and mechanisms underlying these global innate immune responses. Since E. chaffeensis strain-dependent active modulation of host cytokine and chemokine signaling pathways could also underlie variable clinical manifestations in HME patients, some of these highly responsive host genes may serve as molecular markers for assessing HME disease status and the treatment outcome.
Supplementary Material
Acknowledgments
We thank the Histology/Immunohistochemistry Core Lab in the Department of Veterinary Biosciences for research support and Mary Ross for immunohistochemistry technical support. We appreciate Yumi Kumagai for assistance on microscopy.
This work was funded by grant R01AI47885 from the National Institutes of Health.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 10 November 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Andersson, H., B. Hartmanova, R. Kuolee, P. Ryden, W. Conlan, W. Chen, and A. Sjostedt. 2006. Transcriptional profiling of host responses in mouse lungs following aerosol infection with type A Francisella tularensis. J. Med. Microbiol. 55263-271. [DOI] [PubMed] [Google Scholar]
- 2.Baltus, T., P. von Hundelshausen, S. F. Mause, W. Buhre, R. Rossaint, and C. Weber. 2005. Differential and additive effects of platelet-derived chemokines on monocyte arrest on inflamed endothelium under flow conditions. J. Leukoc. Biol. 78435-441. [DOI] [PubMed] [Google Scholar]
- 3.Barnewall, R. E., and Y. Rikihisa. 1994. Abrogation of gamma interferon-induced inhibition of Ehrlichia chaffeensis infection in human monocytes with iron-transferrin. Infect. Immun. 624804-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beurton, F., G. Gueret, M. Horisberger, G. Cheron, and T. Cresteil. 1999. Transcriptional activation of CYP2C, MxA and Fas in sudden infant death syndrome. Int. J. Mol. Med. 333-39. [DOI] [PubMed] [Google Scholar]
- 5.Bluyssen, H. A., R. J. Vlietstra, P. W. Faber, E. M. Smit, A. Hagemeijer, and J. Trapman. 1994. Structure, chromosome localization, and regulation of expression of the interferon-regulated mouse Ifi54/Ifi56 gene family. Genomics 24137-148. [DOI] [PubMed] [Google Scholar]
- 6.Bosma, M. J., and A. M. Carroll. 1991. The SCID mouse mutant: definition, characterization, and potential uses. Annu. Rev. Immunol. 9323-350. [DOI] [PubMed] [Google Scholar]
- 7.Cacalano, G., J. Lee, K. Kikly, A. M. Ryan, S. Pitts-Meek, B. Hultgren, W. I. Wood, and M. W. Moore. 1994. Neutrophil and B cell expansion in mice that lack the murine IL-8 receptor homolog. Science 265682-684. [DOI] [PubMed] [Google Scholar]
- 8.Chang, S. K., B. K. Arendt, J. R. Darce, X. Wu, and D. F. Jelinek. 2006. A role for BLyS in the activation of innate immune cells. Blood 1082687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charo, I. F., and R. M. Ransohoff. 2006. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 354610-621. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, C., C. D. Paddock, and R. Reddy Ganta. 2003. Molecular heterogeneity of Ehrlichia chaffeensis isolates determined by sequence analysis of the 28-kilodalton outer membrane protein genes and other regions of the genome. Infect. Immun. 71187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colotta, F., F. Re, M. Muzio, R. Bertini, N. Polentarutti, M. Sironi, J. G. Giri, S. K. Dower, J. E. Sims, and A. Mantovani. 1993. Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science 261472-475. [DOI] [PubMed] [Google Scholar]
- 12.Colotta, F., S. Saccani, J. G. Giri, S. K. Dower, J. E. Sims, M. Introna, and A. Mantovani. 1996. Regulated expression and release of the IL-1 decoy receptor in human mononuclear phagocytes. J. Immunol. 1562534-2541. [PubMed] [Google Scholar]
- 13.Cyster, J. G. 2003. Lymphoid organ development and cell migration. Immunol. Rev. 1955-14. [DOI] [PubMed] [Google Scholar]
- 14.Czitrom, A. A., S. Edwards, R. A. Phillips, M. J. Bosma, P. Marrack, and J. W. Kappler. 1985. The function of antigen-presenting cells in mice with severe combined immunodeficiency. J. Immunol. 1342276-2280. [PubMed] [Google Scholar]
- 15.Dawson, J. E., B. E. Anderson, D. B. Fishbein, J. L. Sanchez, C. S. Goldsmith, K. H. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 292741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Clerck, L. S., C. M. De Gendt, C. H. Bridts, N. Van Osselaer, and W. J. Stevens. 1995. Expression of neutrophil activation markers and neutrophil adhesion to chondrocytes in rheumatoid arthritis patients: relationship with disease activity. Res. Immunol. 14681-87. [DOI] [PubMed] [Google Scholar]
- 17.Doan, T., R. Melvold, S. Viselli, and C. Waltenbaugh. 2007. Lippincott's illustrated reviews: immunology. Lippincott Williams & Wilkins, Hagerstown, MD.
- 18.Drenkard, D., F. M. Becke, J. Langstein, T. Spruss, L. A. Kunz-Schughart, T. E. Tan, Y. C. Lim, and H. Schwarz. 2007. CD137 is expressed on blood vessel walls at sites of inflammation and enhances monocyte migratory activity. FASEB J. 21456-463. [DOI] [PubMed] [Google Scholar]
- 19.Fichtner-Feigl, S., W. Strober, K. Kawakami, R. K. Puri, and A. Kitani. 2006. IL-13 signaling through the IL-13α2 receptor is involved in induction of TGF-β1 production and fibrosis. Nat. Med. 1299-106. [DOI] [PubMed] [Google Scholar]
- 20.Flo, T. H., K. D. Smith, S. Sato, D. J. Rodriguez, M. A. Holmes, R. K. Strong, S. Akira, and A. Aderem. 2004. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432917-921. [DOI] [PubMed] [Google Scholar]
- 21.Gooptu, B., and D. A. Lomas. 2008. Polymers and inflammation: disease mechanisms of the serpinopathies. J. Exp. Med. 2051529-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruys, E., M. J. Toussaint, T. A. Niewold, and S. J. Koopmans. 2005. Acute phase reaction and acute phase proteins. J. Zhejiang Univ. Sci. B 61045-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hochepied, T., F. G. Berger, H. Baumann, and C. Libert. 2003. Alpha(1)-acid glycoprotein: an acute phase protein with inflammatory and immunomodulating properties. Cytokine Growth Factor Rev. 1425-34. [DOI] [PubMed] [Google Scholar]
- 24.Ismail, N., L. Soong, J. W. McBride, G. Valbuena, J. P. Olano, H. M. Feng, and D. H. Walker. 2004. Overproduction of TNF-alpha by CD8+ type 1 cells and down-regulation of IFN-gamma production by CD4+ Th1 cells contribute to toxic shock-like syndrome in an animal model of fatal monocytotropic ehrlichiosis. J. Immunol. 1721786-1800. [DOI] [PubMed] [Google Scholar]
- 25.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20197-216. [DOI] [PubMed] [Google Scholar]
- 26.Khan, I. A., J. A. MacLean, F. S. Lee, L. Casciotti, E. DeHaan, J. D. Schwartzman, and A. D. Luster. 2000. IP-10 is critical for effector T cell trafficking and host survival in Toxoplasma gondii infection. Immunity 12483-494. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura, A., K. Takahashi, A. Okajima, and N. Kitamura. 1994. Induction of the human gene for p44, a hepatitis-C-associated microtubular aggregate protein, by interferon-alpha/beta. Eur. J. Biochem. 224877-883. [DOI] [PubMed] [Google Scholar]
- 28.Kofler, S., T. Nickel, and M. Weis. 2005. Role of cytokines in cardiovascular diseases: a focus on endothelial responses to inflammation. Clin. Sci. (London) 108205-213. [DOI] [PubMed] [Google Scholar]
- 29.Krishnaswamy, G., J. Kelley, L. Yerra, J. K. Smith, and D. S. Chi. 1999. Human endothelium as a source of multifunctional cytokines: molecular regulation and possible role in human disease. J. Interferon Cytokine Res. 1991-104. [DOI] [PubMed] [Google Scholar]
- 30.Li, X. X., J. H. Huang, H. Y. Rienhoff, Jr., and W. S. Liao. 1990. Two adjacent C/EBP-binding sequences that participate in the cell-specific expression of the mouse serum amyloid A3 gene. Mol. Cell. Biol. 106624-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logsdon, L. K., and J. Mecsas. 2006. A non-invasive quantitative assay to measure murine intestinal inflammation using the neutrophil marker lactoferrin. J. Immunol. Methods 313183-190. [DOI] [PubMed] [Google Scholar]
- 32.Lu, B., B. J. Rutledge, L. Gu, J. Fiorillo, N. W. Lukacs, S. L. Kunkel, R. North, C. Gerard, and B. J. Rollins. 1998. Abnormalities in monocyte recruitment and cytokine expression in monocyte chemoattractant protein 1-deficient mice. J. Exp. Med. 187601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luster, A. D., and J. V. Ravetch. 1987. Genomic characterization of a gamma-interferon-inducible gene (IP-10) and identification of an interferon-inducible hypersensitive site. Mol. Cell. Biol. 73723-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda, K., N. Markowitz, R. C. Hawley, M. Ristic, D. Cox, and J. E. McDade. 1987. Human infection with Ehrlichia canis, a leukocytic rickettsia. N. Engl. J. Med. 316853-856. [DOI] [PubMed] [Google Scholar]
- 35.McQuiston, J. H., C. D. Paddock, R. C. Holman, and J. E. Childs. 1999. The human ehrlichioses in the United States. Emerg. Infect. Dis. 5635-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miura, K., and Y. Rikihisa. 2007. Virulence potential of Ehrlichia chaffeensis strains of distinct genome sequences. Infect. Immun. 753604-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moser, B., and K. Willimann. 2004. Chemokines: role in inflammation and immune surveillance. Ann. Rheum. Dis. 63(Suppl. 2):ii84-ii89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller, A., J. O'Rourke, P. Chu, C. C. Kim, P. Sutton, A. Lee, and S. Falkow. 2003. Protective immunity against Helicobacter is characterized by a unique transcriptional signature. Proc. Natl. Acad. Sci. USA 10012289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller, A., J. O'Rourke, J. Grimm, K. Guillemin, M. F. Dixon, A. Lee, and S. Falkow. 2003. Distinct gene expression profiles characterize the histopathological stages of disease in Helicobacter-induced mucosa-associated lymphoid tissue lymphoma. Proc. Natl. Acad. Sci. USA 1001292-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagalakshmi, M. L., E. Murphy, T. McClanahan, and R. de Waal Malefyt. 2004. Expression patterns of IL-10 ligand and receptor gene families provide leads for biological characterization. Int. Immunopharmacol. 4577-592. [DOI] [PubMed] [Google Scholar]
- 41.Ng, P. C., G. Li, K. M. Chui, W. C. Chu, K. Li, R. P. Wong, K. W. Chik, E. Wong, and T. F. Fok. 2004. Neutrophil CD64 is a sensitive diagnostic marker for early-onset neonatal infection. Pediatr. Res. 56796-803. [DOI] [PubMed] [Google Scholar]
- 42.Ozaki, K., K. Kikly, D. Michalovich, P. R. Young, and W. J. Leonard. 2000. Cloning of a type I cytokine receptor most related to the IL-2 receptor beta chain. Proc. Natl. Acad. Sci. USA 9711439-11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paddock, C. D., and J. E. Childs. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 1637-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pathan, N., C. A. Hemingway, A. A. Alizadeh, A. C. Stephens, J. C. Boldrick, E. E. Oragui, C. McCabe, S. B. Welch, A. Whitney, P. O'Gara, S. Nadel, D. A. Relman, S. E. Harding, and M. Levin. 2004. Role of interleukin 6 in myocardial dysfunction of meningococcal septic shock. Lancet 363203-209. [DOI] [PubMed] [Google Scholar]
- 45.Plougastel, B., K. Matsumoto, C. Dubbelde, and W. M. Yokoyama. 2001. Analysis of a 1-Mb BAC contig overlapping the mouse Nkrp1 cluster of genes: cloning of three new Nkrp1 members, Nkrp1d, Nkrp1e, and Nkrp1f. Immunogenetics 53592-598. [DOI] [PubMed] [Google Scholar]
- 46.Pober, J. S. 2002. Endothelial activation: intracellular signaling pathways. Arthritis Res. 4(Suppl 3)S109-S116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabinowich, H., L. Pricop, R. B. Herberman, and T. L. Whiteside. 1994. Expression and function of CD7 molecule on human natural killer cells. J. Immunol. 152517-526. [PubMed] [Google Scholar]
- 48.Redpath, S., P. Ghazal, and N. R. Gascoigne. 2001. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 986-92. [DOI] [PubMed] [Google Scholar]
- 49.Rikihisa, Y., S. Yamamoto, I. Kwak, Z. Iqbal, G. Kociba, J. Mott, and W. Chichanasiriwithaya. 1994. C-reactive protein and alpha 1-acid glycoprotein levels in dogs infected with Ehrlichia canis. J. Clin. Microbiol. 32912-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roos, D., and R. S. Weening. 1978. Defects in the oxidative killing of microorganisms by phagocytic leukocytes. Ciba Found. Symp. 1978225-262. [PubMed] [Google Scholar]
- 51.Rossi, D., and A. Zlotnik. 2000. The biology of chemokines and their receptors. Annu. Rev. Immunol. 18217-242. [DOI] [PubMed] [Google Scholar]
- 52.Rot, A., and U. H. von Andrian. 2004. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 22891-928. [DOI] [PubMed] [Google Scholar]
- 53.Ruckert, R., S. Bulfone-Paus, and K. Brandt. 2008. Interleukin-21 stimulates antigen uptake, protease activity, survival and induction of CD4+ T cell proliferation by murine macrophages. Clin. Exp. Immunol. 151487-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmekel, B., S. E. Karlsson, M. Linden, C. Sundstrom, H. Tegner, and P. Venge. 1990. Myeloperoxidase in human lung lavage. I. A marker of local neutrophil activity. Inflammation 14447-454. [DOI] [PubMed] [Google Scholar]
- 55.Schwarz, H., F. J. Blanco, J. von Kempis, J. Valbracht, and M. Lotz. 1996. ILA, a member of the human nerve growth factor/tumor necrosis factor receptor family, regulates T-lymphocyte proliferation and survival. Blood 872839-2845. [PubMed] [Google Scholar]
- 56.Sehdev, A. E., and J. S. Dumler. 2003. Hepatic pathology in human monocytic ehrlichiosis. Ehrlichia chaffeensis infection. Am. J. Clin. Pathol. 119859-865. [DOI] [PubMed] [Google Scholar]
- 57.Simonson, G. D., A. C. Vincent, K. J. Roberg, Y. Huang, and V. Iwanij. 1994. Molecular cloning and characterization of a novel liver-specific transport protein. J. Cell Sci. 1071065-1072. [DOI] [PubMed] [Google Scholar]
- 58.Su, G. L. 2002. Lipopolysaccharides in liver injury: molecular mechanisms of Kupffer cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 283G256-G265. [DOI] [PubMed] [Google Scholar]
- 59.Sunil, V. R., K. J. Patel, M. Nilsen-Hamilton, D. E. Heck, J. D. Laskin, and D. L. Laskin. 2007. Acute endotoxemia is associated with upregulation of lipocalin 24p3/Lcn2 in lung and liver. Exp. Mol. Pathol. 83177-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Velayudham, A., I. Hritz, A. Dolganiuc, P. Mandrekar, E. Kurt-Jones, and G. Szabo. 2006. Critical role of toll-like receptors and the common TLR adaptor, MyD88, in induction of granulomas and liver injury. J. Hepatol. 45813-824. [DOI] [PubMed] [Google Scholar]
- 61.von Hundelshausen, P., K. S. Weber, Y. Huo, A. E. Proudfoot, P. J. Nelson, K. Ley, and C. Weber. 2001. RANTES deposition by platelets triggers monocyte arrest on inflamed and atherosclerotic endothelium. Circulation 1031772-1777. [DOI] [PubMed] [Google Scholar]
- 62.Wahl, S. M. 2007. Transforming growth factor-beta: innately bipolar. Curr. Opin. Immunol. 1955-62. [DOI] [PubMed] [Google Scholar]
- 63.Winslow, G. M., E. Yager, K. Shilo, D. N. Collins, and F. K. Chu. 1998. Infection of the laboratory mouse with the intracellular pathogen Ehrlichia chaffeensis. Infect. Immun. 663892-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshiie, K., H. Y. Kim, J. Mott, and Y. Rikihisa. 2000. Intracellular infection by the human granulocytic ehrlichiosis agent inhibits human neutrophil apoptosis. Infect. Immun. 681125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, J. Z., M. Sinha, B. A. Luxon, and X. J. Yu. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect. Immun. 72498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.