Abstract
Mouse-derived macrophages have the unique ability to restrict or permit Legionella pneumophila intracellular growth. The common inbred mouse strain C57BL/6J (B6) restricts L. pneumophila growth, whereas macrophages derived from A/J mice allow >103-fold bacterial growth within three days. This phenotypic difference was mapped to the mouse Naip5 allele. The B6 restrictive Naip5 allele is dominant, and six amino acid changes in its product were predicted to control permissiveness. By using the wild-derived mouse strain MOLF/Ei, we found that MOLF/Ei-derived macrophages also restrict L. pneumophila growth, yet the Naip5 protein is identical to the A/J Naip5 at the six-amino-acid signature. The MOLF/Ei restrictive trait, unlike that of B6-derived macrophages, was not dominant over the A/J trait. In spite of this phenotypic difference, the L. pneumophila growth restriction in MOLF/Ei macrophages was mapped to the Naip5 region as well, indicating that the originally predicted change in the A/J Naip5 allele may not be critical for restriction. In the product of the A/J Naip5 permissive allele, there are four unique amino acid changes that map to a NACHT-like domain. Similar misregulating mutations have been identified in the NACHT domains of Nod-like receptor (NLR) proteins. Therefore, one of these mutations may be critical for restriction of L. pneumophila intracellular growth, and this parallels results found with human NLR variants with defects in the innate immune response.
Legionella pneumophila is a gram-negative intracellular bacterial pathogen. It is found ubiquitously in aquatic environments, where it parasitizes a variety of protozoan species (15). The aerosolization of L. pneumophila from contaminated public water supplies, such as steam baths, cooling towers, or large air conditioning systems, is thought to be the primary route for human infection (17). After it is inhaled, L. pneumophila is able to replicate within alveolar macrophages, resulting in a severe pneumonia known as Legionnaires' disease (17).
Dot/Icm, a chromosomally encoded type IV secretion system, is required for L. pneumophila to replicate within a membrane-bound vacuole in host cells (32, 39). The Legionella-containing vacuole avoids fusion with endosomes and lysosomes (24) and recruits endoplasmic reticulum-derived secretory vesicles that modify the Legionella-containing vacuole into a compartment in which endoplasmic reticulum-like material is imbedded (23, 35, 37). L. pneumophila replicates within macrophages for up to 24 h and then lyses out to repeat the infection cycle.
Macrophages derived from many inbred mouse strains have been shown to be restrictive or permissive of L. pneumophila intracellular growth, with the C57BL/6J (B6) strain being used as the canonical restrictive strain (9, 42, 43). A/J, a permissive mouse strain, supports 103- to 104-fold growth of L. pneumophila over a 3-day period, whereas B6 rarely supports <10-fold growth over this time period (9, 42, 43). Studies involving crosses of B6 and A/J showed that restriction of L. pneumophila growth is dominant and segregates in a Mendelian fashion via a single autosomal locus on mouse chromosome 13 named Lgn1 (5, 9).
The Lgn1 locus contains a variable number of Naip gene paralogs (∼5 genes and pseudogenes) that share ∼85% identity (19, 25). Positional cloning and complementation assays linked L. pneumophila restriction in mouse macrophages to a single gene called Naip5 (also known as Birc1e) (10, 41). Naip5 is a nucleotide-binding domain-containing and leucine-rich Nod-like receptor (NLR) protein made up of three N-terminal baculoviral inhibitory repeat domains, a central NOD/NACHT domain, and C-terminal leucine-rich-repeat motifs (41). NLRs are cytosolic proteins that sense pathogen-associated molecular patterns, common microbial molecules that are released into the cytoplasm of the host cells, often as the result of microbial infection (33). Naip5 is believed to sense L. pneumophila flagellin, dependent on the presence of the type IV secretion system (29, 30). Additional regulation of Legionella infection via phagosome maturation is provided by another NLR protein, Ipaf (NLRC4) (3, 44). The cytoplasmic presentation of flagellin activates caspase-1 and restricts L. pneumophila intracellular growth (3, 16, 29, 30).
The genetic difference distinguishing Naip5 in permissive A/J and restrictive B6 macrophages has been hypothesized to be linked to either the expression level or the amino acid sequence of Naip5, because both strains are predicted to encode an intact Naip5 protein (10, 41). In terms of expression, Diez et al., in 2000, investigated the mRNA expression level of the Naip homologous transcripts in B6 macrophages versus A/J macrophages by Northern blot analysis and showed there was enhanced expression of Naip transcripts in B6 macrophages (11). Wright et al., in 2003, looked at the expression level of Naip5 protein in B6 macrophages versus A/J macrophages and observed enhanced protein expression in B6 macrophages as well (41). However, in both studies, the detection methods did not rule out the possibility that other Naip paralogs could be contributing to these results. Therefore, it is unclear whether the enhanced expression level of Naip in B6-derived macrophages is specific to Naip5.
It was also observed that the Naip5 protein sequence is polymorphic. There are 14 differences in the amino acids encoded by the A/J and B6 Naip5 alleles, suggesting that one of these variants could affect the function of Naip5 (41). A previous study narrowed down the putative amino acids regulating Naip5 activity by evaluating L. pneumophila intracellular growth in seven different inbred mouse strains (41). Through this analysis the authors found two strains (B6 and P/J) that restricted L. pneumophila intracellular growth, and five strains (A/J, C3H/HeJ [C3H], BALB/cJ [BALB], 129S1, and FvB/N) that were permissive (41). The Naip5 gene was sequenced from each strain and six amino acid differences correlated with the permissive/restrictive phenotype (41). These six amino acid changes appeared to serve as a signature for a permissive or restrictive L. pneumophila mouse strain.
To expand upon these studies, we characterized L. pneumophila intracellular growth in the wild-derived mouse strain MOLF/Ei. Although inbred mouse strains have served as a genetic reservoir for pathogenesis, new emerging mouse models, such as wild-derived mice, have expanded the genetic repertoire, allowing novel genes and/or regulatory mechanisms that could play a role in determining host-microbe interactions to be identified (8, 31). We characterized L. pneumophila intracellular growth in MOLF/Ei-derived bone marrow macrophages (BM macrophages) to gain further insight into the genetic determinants regulating L. pneumophila restriction. Surprisingly, we found that MOLF/Ei-derived macrophages restrict L. pneumophila intracellular growth, even though the six-amino-acid signature found in the MOLF/Ei Naip5 allele product is identical to the those encoded by alleles from the permissive FvB/NJ, BALB, and C3H strains. By mapping the L. pneumophila restriction phenotype, we found that it was also linked to Naip5 in MOLF/Ei macrophages, suggesting that the originally proposed missense amino acids in the A/J protein are not critical for L. pneumophila intracellular growth. Instead, we propose that unique amino acid changes within the NACHT domain of A/J Naip5 are likely to be responsible for the permissiveness of this inbred mouse strain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. pneumophila philadelphia-1 strain Lp02 (thyA) is a derivative of Lp01 (hsdR rpsL) (6). The Lp02 flaA-negative strain (referred to as the flaA mutant strain) was generously provided by Tao Ren and William Dietrich (30). L. pneumophila strains were maintained on buffered charcoal-yeast extract solid medium and ACES [N-(2-acetamido)-2-aminoethanesulfonic acid]-buffered yeast extract (AYE) broth culture media (13, 18, 35). For all Lp02 derivatives, thymidine was included in the media at 100 μg/ml. For infections, L. pneumophila strains were patched from a single colony onto buffered charcoal-yeast extract containing 100 μg/ml of thymidine. After 2 days at 37°C, patches were used to inoculate AYE broth cultures. Cultures were grown overnight in AYE broth containing 100 μg/ml of thymidine to ensure that the bacteria were in postexponential phase (A600, ∼4.0) prior to infection.
Mice, phenotyping, and genotyping.
A/J, B6, MOLF/Ei, and FvB/NJ were purchased from Jackson Laboratory. A/J and MOLF/Ei were crossed to generate F1 progeny. The F1 progeny were backcrossed to A/J, and N2 (F1 × A/J) progeny were phenotyped using the L. pneumophila growth curve assay (see below). Legionella growth was assessed at 2, 48, and 72 h postinfection (hpi), and mice were scored as either intermediate (102-fold growth) or permissive (103-fold growth) for L. pneumophila growth in comparison with an A/J or (MOLF/Ei × A/J)F1 control. Genotyping of N2 (F1 × A/J) progeny was performed with polymorphic microsatellite PCR using genomic DNA obtained from tail tissue (Qiagen).
Cell culture.
BM macrophages were flushed from the femurs of 6-week-old to 3-month-old mice and differentiated in BM macrophage medium (BMM; RPMI, 1 mM glutamine, 10% fetal bovine serum, 30% L-cell supernatant) (35). Macrophages were differentiated for 7 to 8 days, collected, and frozen for use in multiple experiments, if needed, in media containing 20% serum and 10% dimethyl sulfoxide (DMSO).
Growth curves for Legionella pneumophila in BM macrophages.
Macrophages were replated after 7 to 8 days of differentiation in fresh BMM plus 200 μg/ml of thymidine. BM macrophages were plated at 4 × 105 cells per well of a 24-well plate and allowed to settle overnight. Legionella pneumophila was grown in AYE broth to postexponential phase (A600, ∼4.0), when the bacteria are highly motile, and the cells were infected at a multiplicity of infection (MOI) of 0.05. After infection of the cells, culture plates were placed in a tabletop centrifuge and spun at 1,000 rpm for 5 min at room temperature to promote contact of bacteria with the macrophages. The macrophages were incubated at 37°C with 5% CO2 for 2 h, after which the monolayers were washed three times in prewarmed BMM plus 200 μg/ml of thymidine. At 2, 24, 48, and 72 hpi, three independent wells at each time point were lysed with 0.2% saponin. Dilutions of each lysate were plated onto bacteriological media, and CFU were determined. For each time point, we determined the mean number of bacteria recovered from three independent wells ± the standard error.
Caspase-1 inhibition.
The specific caspase-1 inhibitor Z-YVAD-FMK (YVAD; Calbiochem) was used. Macrophages were preincubated 1 h before infection with 40 μM of inhibitor dissolved in DMSO or with an equivalent volume of DMSO (control). Macrophages were infected as described above, and after the third wash, fresh inhibitor was added for the duration of growth.
Cloning and sequencing Naip5 from MOLF/Ei BMM.
Total RNA was isolated from MOLF/Ei BM macrophages according to the instructions with the Qiagen RNeasy kit (Invitrogen). cDNA was amplified from 5 to 10 μg of total RNA by using oligo(dT) and Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Naip5 was amplified from cDNA by using Pfu Ultra DNA polymerase (Stratagene) with primers Naip5F-myc and Naip5R and cloned into pMyc (Clontech) at EcoRI and SalI sites. pMyc-Naip5 clones were screened by restriction digested and sequenced using primers Naip5F-myc and Naip5R (Table 1). The full-length sequence of Naip5 was determined using the primers Naip5FS1-6 and Naip5RS1-6 (Table 1). HEK293T cells were transfected with MOLF Naip5 cDNA, and lysates were prepared for Western blotting to confirm expression of the full-length protein (data not shown).
TABLE 1.
List of primers
| Primer | Restriction site | Sequencea |
|---|---|---|
| Naip5F-myc | EcoRI | CCGGAATTCGGATGGCTGAGCATGGGGAG |
| Naip5FS1 | GGTACCATGAAGAGGAGG | |
| Naip5FS2 | GTAGGAGTGAAGCCCAG | |
| Naip5FS3 | CTTCTATAATACTGTCTC | |
| Naip5FS4 | GTTTCAGTTTGTTAGAGG | |
| Naip5FS5 | CATGTCCAGGCTGGAGCT | |
| Naip5FS6 | CTGCAGCTTCCGTGCCTC | |
| Naip5FS7 | GAAGCTCTAGTCAGAGCAGG | |
| Naip5RS1 | CCTCCTCTTCATGGTACC | |
| Naip5RS2 | ACTGGGCTTCACTCCTAC | |
| Naip5RS3 | GAGACAGTATTATAGAAG | |
| Naip5RS4 | CCTCTAACAAACTGAAAC | |
| Naip5RS5 | AGCTCCAGCCTGGACATG | |
| Naip5RS6 | GAGGCACGGAAGCTGCAG | |
| Naip5R | SalI | ACGCGTCGACCCAGGAGGGCCCAACATAC |
Restriction sites are indicated in boldface.
RNA isolation and real-time PCR.
Cells were lysed for RNA preparation using 1 ml of Trizol (Invitrogen) per 106 cells. The RNA pellet was reconstituted in 20 μl of water and treated with 20 U of DNase Q1 (Promega) for 1 h at 37C. RNA was extracted with phenol and phenol-chloroform, reprecipitated, and subsequently used for cDNA synthesis using random primers of nine nucleotides, deoxynucleoside triphosphates, and Moloney murine leukemia virus reverse transcriptase (New England Biolabs). cDNA was analyzed by real-time PCR gene expression analysis using the Sybr green PCR master mix kit (Applied Biosystems) and the Naip5-specific primers Naip5F (5′-GTG CTG GTC ACC AAA CCT TTA TC-3′), Nip5R (5′-TCC TGT TGA CCT TGG TAT TGG AAG-3′), GAPDH_F (5′-CCA TGG AGA AGG CTG GGG), and GAPDH_R (5′-CAA AGT TGT CAT GGA TGA CC).
Data analysis.
An enhanced version of Map Manager QT (quantitative traits) software, QTX, was used in linkage analysis of the growth permissiveness/restriction trait. Linkages with a logarithm of odds (LOD) greater than 3.0 were considered significant. All experiments were performed in triplicate unless stated otherwise. Statistical analysis of differences in expression of Naip5 was carried out using two-sample t test.
RESULTS
MOLF/Ei-derived BM macrophages restrict L. pneumophila growth.
Wild-derived mice, such as MOLF/Ei strain mice, have many advantages over the commonly used inbred mouse strains, including novel allelic variants and greater density of single-nucleotide polymorphisms, that facilitate mapping strategies (20). Most of the commonly used inbred mouse strains have been established from the Mus musculus domesticus subspecies, representing a relatively small gene pool, making these strains limited in genetic variations. In contrast, wild-derived mice originated mainly from M. m. musculus and M. m. castaneus subspecies are genetically diverse compared to the classical inbred strains. Compared to one another, the genomes of the wild-derived and classical inbred strains show a polymorphism in every 100 to 200 bp (34), suggesting that screening wild-derived mice may uncover other host factors or novel alleles of previously characterized genes that influence L. pneumophila intracellular growth.
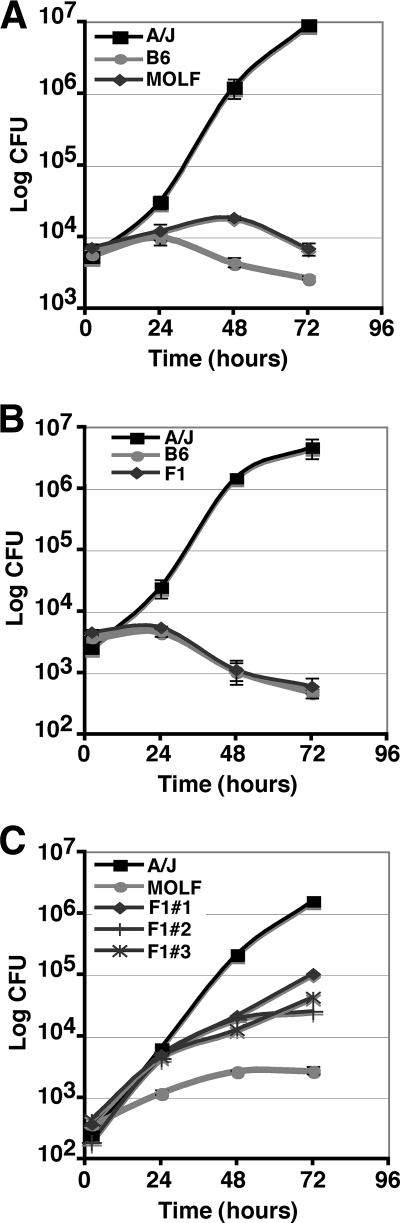
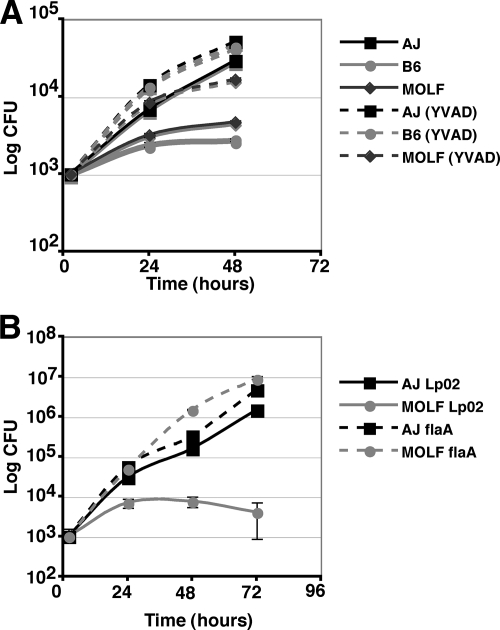
To address this point, L. pneumophila intracellular growth was assayed in BM macrophages over a 3-day period. BM macrophages from the A/J strain, the B6 strain, and the wild-derived mouse strain MOLF/Ei were infected with Lp02 at a low dose (MOI, 0.05), and L. pneumophila growth was monitored by enumeration of CFU at 2, 24, 48, and 72 hpi. We found that MOLF/Ei BM macrophages strongly restricted Lp02 growth to an extent similar to that observed for the commonly used restrictive inbred B6 mouse strain (Fig. 1A).
FIG. 1.
MOLF/Ei-derived macrophages restrict L. pneumophila growth. Growth curves for L. pneumophila (Lp02) in A/J, MOLF/Ei, and B6 strains (A), A/J, B6, and (A/J × B6)F1 strains (B), and A/J, MOLF/Ei, and (MOLF/Ei × A/J)F1 strains (C). Three independent F1 mice were tested and labeled arbitrarily numbers 1 to 3. For each growth curve experiment, BM macrophages were infected with Lp02 at an MOI of 0.05. Cells were lysed at 2, 24, 48, and 72 hpi, and CFU were enumerated. Data represent the means and standard errors for triplicate samples.
Previous work identified six amino acid changes within Naip5 that were present in all strains of mice that were permissive of L. pneumophila intracellular growth (Table 2) (41). Naip5 was cloned and sequenced from cDNA prepared from MOLF/Ei-derived macrophages to determine whether the L. pneumophila restriction in MOLF/Ei macrophages could be due to mutations in Naip5. The signature six polymorphic amino acids in the MOLF/Ei Naip5 protein were identical to those in the A/J protein, yet MOLF/Ei-derived macrophages were restrictive for L. pneumophila growth (Table 2).
TABLE 2.
Naip5 amino acid polymorphisms in B6, MOLF, and A/J mice
| Amino acid no. | Exon | Polymorphic residuea
|
||
|---|---|---|---|---|
| B6 | MOLF | A/J | ||
| 92 | 3 | R | R | R |
| 144 | 3 | R | R | R |
| 234 | 5 | E | K | E |
| 368 | 9 | T | M | T |
| 472 | 11 | T | A | A |
| 496 | 11 | Y | Y | N |
| 512 | 11 | D | D | G |
| 514 | 11 | G | G | E |
| 517 | 11 | N | N | K |
| 533 | 11 | V | A | A |
| 538 | 11 | S | I | I |
| 647 | 11 | A | A | T |
| 692 | 11 | S | S | P |
| 755 | 11 | V | V | M |
| 855 | 11 | S | T | S |
| 952 | 11 | S | T | S |
| 1021 | 11 | M | I | M |
| 1092 | 12 | E | D | D |
| 1116 | 12 | N | D | D |
| 1123 | 12 | R | G | G |
| 1137 | 12 | Q | R | Q |
| 1140 | 12 | T | R | T |
| 1241 | 15 | V | V | I |
| 1275 | 15 | D | D | N |
Naip5 amino acids in bold were originally predicted to correlate with permissiveness of mouse macrophages for L. pneumophila growth (41). Residues which are unique to A/J Naip5 compared to those of B6 or MOLF Naip5 are underlined.
The B6 restrictive Naip5 allele is known to be dominant over the A/J sensitive Naip5 allele. Consistent with this observation, we found that BM macrophages from (B6 × A/J)F1 progeny restricted L. pneumophila growth (Fig. 1B). BM macrophages from (MOLF/Ei × A/J)F1 were tested to determine whether the MOLF/Ei Naip5 allele was dominant or recessive. Surprisingly, all seven F1 crosses (MOLF/Ei × A/J) tested in our study were intermediate for L. pneumophila intracellular growth (Fig. 1C; data not shown). We consistently observed a 10-fold defect in L. pneumophila growth when the (MOLF/Ei × A/J)F1 strain was compared to the A/J strain. Altogether, the sequence similarity to a permissive Naip5 allele and the difference in the (MOLF/Ei × A/J)F1 mode of inheritance of this trait indicated that another gene(s) beside Naip5 may be influencing the restrictive MOLF/Ei phenotype. Alternatively, the restrictive B6 Naip5 allele may behave differently than a restrictive MOLF/Ei allele.
L. pneumophila restriction in MOLF/Ei-derived macrophages is linked to Naip5.
In contrast to the B6 Naip5 allele, the MOLF/Ei Naip5 allele was not dominant over the A/J Naip5 allele, so we hypothesized that another gene(s) within the MOLF/Ei strain background may be contributing to the restriction of L. pneumophila growth. To test this, the progeny of (MOLF/Ei × A/J)F1 mice were backcrossed to strain A/J to map the genetic difference influencing the L. pneumophila phenotype.
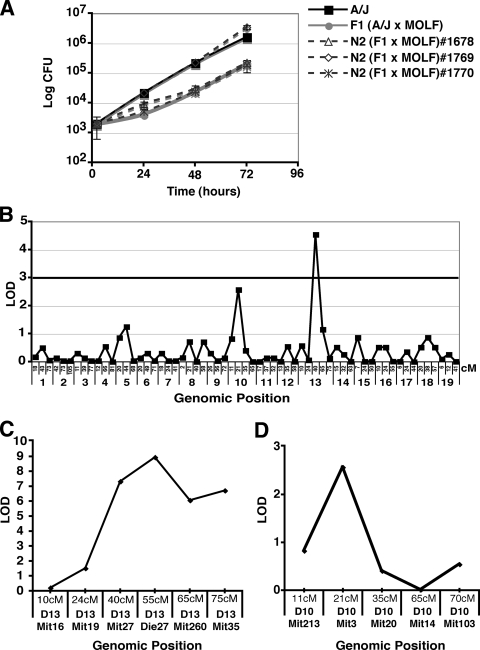
BM macrophages from the resulting N2 (F1 × A/J) mice were screened for phenotypic differences in L. pneumophila growth. A total of 75 N2 (F1 × A/J) mice were screened by assaying for L. pneumophila growth in BM macrophages at 2, 48, and 72 hpi. Of the 75 mice screened, 53% of N2 (F1 × A/J) mice were permissive (A/J-like) and 47% were found to be intermediate (F1-like) for L. pneumophila growth, suggesting a simple Mendelian segregation pattern and suggesting that a single gene may regulate the restriction phenotype (Fig. 2A; data not shown). Genome-wide scanning was performed on 28 N2 mice by using 62 polymorphic microsatellite markers spaced at 20- to 30-centimorgan (cM) intervals on each chromosome. The strongest linkage observed was to chromosome 13 at 40 cM, with a transgenomic log likelihood (LOD) of 4.54 (Fig. 2B). Further evaluation of linkage to chromosome 13 using 47 additional mice revealed that the strongest linkage was to marker D13Die27, located within the intergenic region between the Naip2 and Naip5 genes (Fig. 2C) (19). The LOD score peaked at D13Die27 (LOD = 8.9). Equivalent linkage was also observed with D13Die35 located within the Naip5 intron (data not shown). We also observed a peak in LOD score at chromosome 10, but the LOD score was less than 3 and failed to increase when the additional 47 mice were included in the analysis, so the linkage was not considered significant (Fig. 2D). Our linkage analysis demonstrated that the primary restriction of L. pneumophila growth in MOLF/Ei macrophages is likely dependent on Naip5.
FIG. 2.
L. pneumophila restriction in MOLF/Ei-derived macrophages maps to Naip5. (A) BM macrophages from N2 mice generated by mating strain (MOLF/Ei × A/J)F1 to strain A/J were tested for permissiveness to L. pneumophila growth. Macrophages from A/J, (MOLF/Ei × A/J)F1, and three representative N2 mice were infected at an MOI of 0.05, cells were lysed, and CFU were enumerated at 2, 48, and 72 hpi. Data represent the means and standard errors for triplicate samples. (B) Genome-wide scanning was performed according to standard procedures, using 62 polymorphic microsatellite markers throughout the genome, spaced at approximately 20- to 30-cM intervals per chromosome. The position of each genetic marker is indicated on the x axis in cM. Transgenomic log likelihood (LOD score) analysis was performed for 28 mice. The bold horizontal line indicates the cutoff for a significant LOD (≥3). (C) Forty-seven N2 mice were subjected to phenotypic analysis and genotyped for six chromosome 13 markers; names and positions (cM) are indicated on the x axis. D13Die27 markers the physical position of the Naip5 allele (19). (D) Forty-seven N2 mice were subjected to phenotypic analysis and genotyped for five chromosome 10 markers; names and positions (cM) are indicated on the x axis.
BM macrophages from FvB/N are not permissive of L. pneumophila intracellular growth.
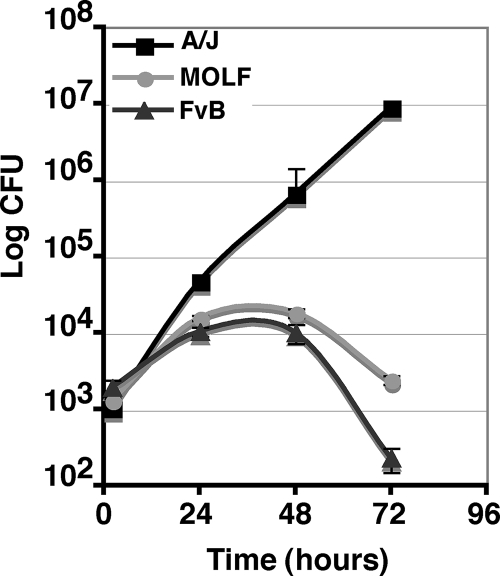
The mapping of L. pneumophila restriction to Naip5 in the MOLF/Ei strain effectively argues that any common amino acid polymorphisms that control permissiveness for L. pneumophila intracellular growth have yet to be identified (Table 2). In fact, the MOLF/Ei Naip5 amino acid sequence is identical to those of the proteins from inbred FvB/NJ, BALB, and C3H mouse strains (data not shown), which were shown to be permissive (41). To evaluate this discrepancy, we performed growth curve analyses with FvB/NJ BM macrophages and found that, in contrast to previous published results, FvB/NJ BM macrophages were restrictive of L. pneumophila intracellular growth (Fig. 3). In support of our data, another study showed that peritoneal macrophages from FvB/NJ mice were also restrictive for L. pneumophila growth (10). Differences in our BM macrophage differentiation or growth curve analysis protocols may also have influenced the phenotype of FvB/NJ. If the identified residues in Naip5 are playing a role in controlling the restrictive/permissive phenotype, then we propose that the critical amino acids are likely the ones that are unique to the A/J Naip5 allele product and are not found in any of the restrictive strains (Table 2).
FIG. 3.
FvB/NJ-derived macrophages restrict L. pneumophila intracellular growth. Growth curve for Lp02 in A/J, MOLF/Ei, and FvB/NJ strains. BM macrophages were infected at an MOI of 0.05, and then cells were lysed at 2, 24, 48, and 72 hpi. Data represent the means and standard errors for triplicate samples.
Unique residue variants in the A/J Naip5 allele product map to the NACHT domain.
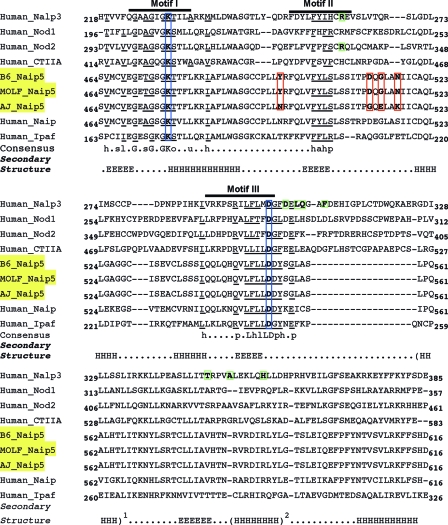
There are nine residues unique to the A/J Naip5 in comparison to the amino acid sequences of the B6 and MOLF/Ei Naip5 proteins (Table 2). Interestingly, four of the unique A/J residues cluster within exon 11, which contains the NACHT nucleoside triphosphatase (NTPase) domain between amino acids 464 and 618. NACHT-containing proteins in mammalian cells are thought to be involved in the innate immune response and the sensing of microbial ligands (40). Using PSI-BLAST and CLUSTALW, we aligned the Naip5 NACHT domains from B6, MOLF/Ei, and A/J Naip5 with the NACHT domains from the human Nalp3, Nod1, Nod2, CTIIA, Naip, and Ipaf proteins (Fig. 4). There are five conserved motifs that are found in these NACHT NTPase proteins (4). Motifs I and III are found in nearly all NTPases, and these motifs contain Walker A and B nucleotide-binding signatures. The Walker A box in motif I has the phosphate-binding site (P-loop), and the Walker B box in motif III has one to three aspartate or glutamate residues, which act to coordinate the Mg2+-water molecule and provide the catalytic carboxylate for NTP hydrolysis (21). Motifs II, IV, and V are present in some, but not all, NACHT NTPases (4). Only motifs I, II, and III were easily discernible in Naip5 (Fig. 4).
FIG. 4.
Alignment of Naip5 to other NACHT domain-containing proteins. Three classic motifs are found in NACHT domain-containing proteins (4). Motif I contains the phosphate-binding lysine (K) of the Walker A box. Motif II contains hydrophobic and conserved polar residues. Motif III contains an aspartate (D) residue to coordinate the Mg2+ of the Walker B motif. NACHT domains from Naip5 of B6, MOLF/Ei, and A/J mouse strains were compared to human Nalp3, CIITA, Nod1, Nod2, Naip, and Ipaf proteins. The consensus amino sequence is present in 90% of NTPases, as described by Aravind et al. in 1999 (4). The amino acid residues within these motifs are underlined and marked as aromatic (a), hydrophobic (h), aliphatic (l), small (s), tiny (u), hydroxyl (o), and polar (p). The conserved Walker box residues are boxed in blue. The sites of the Nalp3 (R260W, D303N, L305P, Q306L, F309S, T348M, A352V, and H358R) and Nod2 (R334W/Q) gain-of-function mutations are shown boxed in green (38). The Naip5 allelic variants Y496N, D512G, G514E, and N517K are shown boxed in red. Marked below each region is the predicted secondary structure from PSIPRED (27). Residues with a score of ≥7 are marked as H for α-helices and E for β-sheets. A superscript 1 indicates that the secondary structure was present only in mouse Naip5s and human Naip and Ipaf. A superscript 2 indicates that the secondary structure was present only in human Naip and Ipaf.
Of particular interest were four amino acids that differ between restrictive B6 and MOLF/Ei Naip5 proteins and the permissive A/J Naip5 protein that cluster near motif II (Fig. 4). This is where other NACHT domain mutations in Nod2 and Nalp3 which result in a gain of function leading to autoinflammatory diseases have been identified (Fig. 4) (1, 2, 12, 7, 14, 22, 38). Protein modeling has mapped these mutations to a loop region near the NTPase active site (2). These mutations could disturb NTP binding and hydrolysis or interfere with protein domain interactions (2). Three of the four amino acid variants in A/J Naip5 are dramatic. The B6 and MOLF/Ei Y496 residue changes from an aromatic residue into a polar residue in the A/J protein. The B6 and MOLF/Ei D512 changes from a negatively charged amino acid into a nonpolar residue and G514 changes from a nonpolar residue into a negatively charged residue in the A/J Naip5 protein (Fig. 4). Based on this model, we propose that the missense amino acids surrounding motif II may be critical for either Naip5 activation or function and may explain why the A/J Naip5 is a permissive allele.
Expression analysis of Naip5 in different mouse strains.
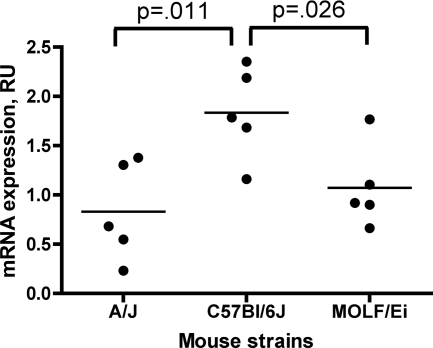
To investigate whether the differences in expression of Naip5 may still explain the phenotypes associated with the different alleles of the gene, we compared the expression levels of Naip5 in B6, A/J, and MOLF/Ei BM macrophages by using a set of primers designed for a region within the 3′ untranscribed region that was specific to Naip5 (Fig. 5). No significant difference was observed between the Naip5 expression levels of A/J and MOLF/Ei macrophages, further supporting the model in which phenotypic differences between these strains are unlikely due to differences in expression levels. In contrast, expression of Naip5 in B6 mice was found to be significantly higher than that in A/J and MOLF/Ei strains (Fig. 5). Therefore, restriction of L. pneumophila growth by mice bearing the B6 allele could be due to high levels of Naip5 in this strain compared to those in the A/J strain.
FIG. 5.
Differences in Naip5 expression in inbred strain macrophages. Isolated from the BM macrophages of A/J, B6, and MOLF/Ei mice, total RNA was analyzed by means of a quantitative PCR that specifically amplifies Naip5. Five mice per strain were used. Significance in variances was evaluated with a one-way analysis of variance test with a 95% confidence interval. RU, relative units.
MOLF/Ei Naip5 restricts L. pneumophila growth by recognition of flagellin and activation of caspase-1.
Naip5 has been shown to be present in a signaling pathway that involves caspase-1 and Ipaf, two members of the inflammasome complex (3, 30, 44). The inflammosome regulates a pyroptosis pathway, which can result in caspase-1-dependent cell death (26). Since knockout of caspase-1 in B6 macrophages can rescue L. pneumophila growth, we wanted to determine whether caspase-1 was critical for MOLF/Ei restriction of L. pneumophila as well. A/J, B6, or MOLF/Ei BM macrophages were treated with or without the membrane-permeable caspase-1 inhibitor YVAD. Consistent with previous findings, wild-type L. pneumophila (Lp02) growth was enhanced ∼10-fold in the B6 strain at 48 hpi (44). Similarly, treatment of MOLF/Ei macrophages with YVAD restored intracellular L. pneumophila growth to the level of that in untreated A/J macrophages at 48 hpi (Fig. 6A). Therefore, restriction of L. pneumophila intracellular growth by macrophages bearing the MOLF/Ei Naip5 allele appeared to occur via the same mechanism as occurs in B6 macrophages, as caspase-1 activation is a critical component in the restriction phenotype observed in both mouse strains.
FIG. 6.
The absence of flagellin or inhibition of caspase-1 restores L. pneumophila growth within MOLF/Ei-derived macrophages. (A) BM macrophages were pretreated for 30 min with DMSO (control) or the caspase-1 inhibitor YVAD. Macrophages from the A/J, B6, and MOLF/Ei strains were infected with Lp02 and assayed for L. pneumophila intracellular growth at 2, 24, and 48 hpi. (B) Growth curve for BM macrophages from A/J or MOLF/Ei mice that were infected with the L. pneumophila Lp02 strain or the flaA mutant strain. Data represent the means and standard errors for triplicate samples.
L. pneumophila flaA mutants, lacking flagellin, fail to activate the Naip5/Ipaf-dependent response that restricts L. pneumophila growth in B6 macrophages (3, 29, 30). It was predicted that flagellin is delivered into the cytosol of mammalian cells, dependent on the presence of Dot/Icm, and is recognized by Naip5, resulting in caspase-1-dependent restriction. Growth curve analyses were conducted using the wild type (Lp02) and a flaA mutant strain, to determine if delivery of L. pneumophila flagellin was contributing to MOLF/Ei restriction. The L. pneumophila flaA mutant strain was able to grow efficiently in B6 (data not shown) as well as MOLF/Ei BM (Fig. 6B) macrophages. We also found that the absence of flagellin was able to fully restore L. pneumophila growth in FvB/NJ BM macrophages (data not shown). This result further supports the model in which L. pneumophila restriction in MOLF/Ei-derived macrophages is due to a restrictive Naip5 allele. The fact that FvB/NJ mouse strain has a Naip5 allele identical to that of MOLF/Ei indicates that several restrictive alleles of Naip5 may operate via recognition of flagellin and activation of caspase-1.
DISCUSSION
Here, we showed that a Naip5-linked determinant controls L. pneumophila growth in the wild-derived mouse strain MOLF/Ei in addition to the commonly used inbred B6 mouse strain. In contrast to previous studies using inbred mice, intriguingly, we observed that the restrictive MOLF/Ei Naip5 allele is not dominant over A/J Naip5. The reason for the different behaviors of the (MOLF/Ei × A/J)F1 and (B6 × A/J)F1 mice is not clear, although the relative expression levels of Naip5 in the MOLF/Ei and B6 strains offer a possible explanation. Naip5 is more highly expressed in the B6 strain than in the MOLF/Ei strain, consistent with the dominant phenotype observed in mice bearing the B6 allele.
One question that still remains is whether the amino acid polymorphisms are critical to the function of Naip5. A study by Zamboni et al. used an HEK293 ectopic expression system to evaluate whether Naip5 was sufficient to induce caspase-1-dependent cell death (44). In that study, HEK293 cells were cotransfected with Fc receptor to increase L. pneumophila uptake and red fluorescent protein to monitor cell morphological changes associated with cell death, caspase-1, and Naip5 from either B6 or A/J mice. The authors showed that upon L. pneumophila infection, only cells expressing the B6 Naip5 allele were sufficient to induce cell death, and there was little cell death with cells expressing the A/J Naip5 allele (44). This result supports a model in which the A/J Naip5 protein is deficient for signaling. We have attempted to determine if the MOLF/Ei allele of Naip5 is able to activate caspase-1 signaling in response to L. pneumophila challenge in HEK293 cells. However, we found that it is difficult to produce a robust caspase-1 response even in the presence of the B6 allele, so we have been unable to test this model directly (data not shown).
To further support a role of the amino acid variants identified in controlling the response to L. pneumophila, similar changes have been identified in members of the NLR family that function in the innate immune response to microbial infection in mammalian cells. We propose that the key residues in A/J Naip5 that lead to inactivation of protein function are clustered near motif II in the NACHT domain (Fig. 4), since other misregulating mutations have been identified in this region. In particular, mutations that contribute to autoinflammatory human diseases have been identified in the NACHT domain of NLR proteins. For instance, several mutations in the NACHT domain of NALP3 (also known as CIAS1, PYPAF1, or cryoprin) have been identified (1, 12, 14, 22). Missense mutations in NALP3 in this region are believed to result in autoactivation leading to the Muckle-Wells syndrome and familial cold urticaria (38), which are associated with an excess of interleukin-1β production. Three mutations in the NACHT domain of Nod2 (R334Q/W and L469F) are associated with Blau syndrome, an autosomal dominant trait leading to granulomatous arthritis, iritis, and skin rash (28), which may result in enhanced activation of transcription factor NF-κB (36). In addition, mutagenesis on NACHT family members has revealed that subtle amino acid changes in this region can affect protein function (38). Transgenic mice carrying mutations affecting these residues in the B6 Naip5 allele product should help to determine if these residues are critical to Naip5 function as well.
In conclusion, the results of our study is consistent with the model in which Naip5 controls L. pneumophila restriction even in the more genetically diverse wild mouse strain, MOLF/Ei. Naip5 restriction is dependent on caspase-1 activation and expression of L. pneumophila flagellin, similar to the more commonly used restrictive B6 strain. However, our study revises the critical amino acid residues in Naip5 function and suggests further examination of the polymorphisms within the NACHT domain. Lastly, the MOLF/Ei Naip5 allele is hemidominant over the A/J Naip5 allele, suggesting that there should be a more complex level of Naip5 regulation in murine macrophages than previously described.
Acknowledgments
We are thankful to Joyce Yang for her helpful comments on the manuscript and Tao Ren for generously supplying the Lp02 flaA mutant strain used in this study.
This work was supported by funding from the Howard Hughes Medical Institute (to R.I.), NIH/NIAID training grant 2T32AI007422, and NIH grant ROAI056234 (to A.P.).
We declare that no competing financial interests exist.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 3 November 2008.
REFERENCES
- 1.Aganna, E., F. Martinon, P. N. Hawkins, J. B. Ross, D. C. Swan, D. R. Booth, H. J. Lachmann, A. Bybee, R. Gaudet, P. Woo, C. Feighery, F. E. Cotter, M. Thome, G. A. Hitman, J. Tschopp, and M. F. McDermott. 2002. Association of mutations in the NALP3/CIAS1/PYPAF1 gene with a broad phenotype including recurrent fever, cold sensitivity, sensorineural deafness, and AA amyloidosis. Arthritis Rheum. 462445-2452. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, M., T. Lengauer, and S. Schreiber. 2003. Disease-associated variants in PYPAF1 and NOD2 result in similar alterations of conserved sequence. Bioinformatics 192171-2175. [DOI] [PubMed] [Google Scholar]
- 3.Amer, A., L. Franchi, T. D. Kanneganti, M. Body-Malapel, N. Ozoren, G. Brady, S. Meshinchi, R. Jagirdar, A. Gewirtz, S. Akira, and G. Nunez. 2006. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 28135217-35223. [DOI] [PubMed] [Google Scholar]
- 4.Aravind, L., V. M. Dixit, and E. V. Koonin. 1999. The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci. 2447-53. [DOI] [PubMed] [Google Scholar]
- 5.Beckers, M. C., S. Yoshida, K. Morgan, E. Skamene, and P. Gros. 1995. Natural resistance to infection with Legionella pneumophila: chromosomal localization of the Lgn1 susceptibility gene. Mamm. Genome 6540-545. [DOI] [PubMed] [Google Scholar]
- 6.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 77-19. [DOI] [PubMed] [Google Scholar]
- 7.Chamaillard, M., D. Philpott, S. E. Girardin, H. Zouali, S. Lesage, F. Chareyre, T. H. Bui, M. Giovannini, U. Zaehringer, V. Penard-Lacronique, P. J. Sansonetti, J. P. Hugot, and G. Thomas. 2003. Gene-environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc. Natl. Acad. Sci. USA 1003455-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conner, J. R., I. I. Smirnova, and A. Poltorak. 2008. Forward genetic analysis of Toll-like receptor responses in wild-derived mice reveals a novel antiinflammatory role for IRAK1BP1. J. Exp. Med. 205305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietrich, W. F., D. M. Damron, R. R. Isberg, E. S. Lander, and M. S. Swanson. 1995. Lgn1, a gene that determines susceptibility to Legionella pneumophila, maps to mouse chromosome 13. Genomics 26443-450. [DOI] [PubMed] [Google Scholar]
- 10.Diez, E., S. H. Lee, S. Gauthier, Z. Yaraghi, M. Tremblay, S. Vidal, and P. Gros. 2003. Birc1e is the gene within the Lgn1 locus associated with resistance to Legionella pneumophila. Nat. Genet. 3355-60. [DOI] [PubMed] [Google Scholar]
- 11.Diez, E., Z. Yaraghi, A. MacKenzie, and P. Gros. 2000. The neuronal apoptosis inhibitory protein (Naip) is expressed in macrophages and is modulated after phagocytosis and during intracellular infection with Legionella pneumophila. J. Immunol. 1641470-1477. [DOI] [PubMed] [Google Scholar]
- 12.Dodé, C., N. Le Du, L. Cuisset, F. Letourneur, J. M. Berthelot, G. Vaudour, A. Meyrier, R. A. Watts, D. G. Scott, A. Nicholls, B. Granel, C. Frances, F. Garcier, P. Edery, S. Boulinguez, J. P. Domergues, M. Delpech, and G. Grateau. 2002. New mutations of CIAS1 that are responsible for Muckle-Wells syndrome and familial cold urticaria: a novel mutation underlies both syndromes. Am. J. Hum. Genet. 701498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feeley, J. C., R. J. Gibson, G. W. Gorman, N. C. Langford, J. K. Rasheed, D. C. Mackel, and W. B. Baine. 1979. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J. Clin. Microbiol. 10437-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldmann, J., A. M. Prieur, P. Quartier, P. Berquin, S. Certain, E. Cortis, D. Teillac-Hamel, A. Fischer, and G. de Saint Basile. 2002. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am. J. Hum. Genet. 71198-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4286-290. [DOI] [PubMed] [Google Scholar]
- 16.Fortier, A., C. de Chastellier, S. Balor, and P. Gros. 2007. Birc1e/Naip5 rapidly antagonizes modulation of phagosome maturation by Legionella pneumophila. Cell. Microbiol. 9910-923. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, D. W. 2005. The challenges were legion. Lancet Infect. Dis. 5237-241. [DOI] [PubMed] [Google Scholar]
- 18.Gabay, J. E., M. Blake, W. D. Niles, and M. A. Horwitz. 1985. Purification of Legionella pneumophila major outer membrane protein and demonstration that it is a porin. J. Bacteriol. 16285-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Growney, J. D., and W. F. Dietrich. 2000. High-resolution genetic and physical map of the Lgn1 interval in C57BL/6J implicates Naip2 or Naip5 in Legionella pneumophila pathogenesis. Genome Res. 101158-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guénet, J. L., and F. Bonhomme. 2003. Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet. 1924-31. [DOI] [PubMed] [Google Scholar]
- 21.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6519-529. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman, H. M., J. L. Mueller, D. H. Broide, A. A. Wanderer, and R. D. Kolodner. 2001. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 1581319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 1582108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, S., J. M. Scharf, J. D. Growney, M. G. Endrizzi, and W. F. Dietrich. 1999. The mouse Naip gene cluster on chromosome 13 encodes several distinct functional transcripts. Mamm. Genome 101032-1035. [DOI] [PubMed] [Google Scholar]
- 26.Lamkanfi, M., T. D. Kanneganti, L. Franchi, and G. Nunez. 2007. Caspase-1 inflammasomes in infection and inflammation. J. Leukoc. Biol. 82220-225. [DOI] [PubMed] [Google Scholar]
- 27.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16404-405. [DOI] [PubMed] [Google Scholar]
- 28.Miceli-Richard, C., S. Lesage, M. Rybojad, A. M. Prieur, S. Manouvrier-Hanu, R. Hafner, M. Chamaillard, H. Zouali, G. Thomas, and J. P. Hugot. 2001. CARD15 mutations in Blau syndrome. Nat. Genet. 2919-20. [DOI] [PubMed] [Google Scholar]
- 29.Molofsky, A. B., B. G. Byrne, N. N. Whitfield, C. A. Madigan, E. T. Fuse, K. Tateda, and M. S. Swanson. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 2031093-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren, T., D. S. Zamboni, C. R. Roy, W. F. Dietrich, and R. E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sancho-Shimizu, V., and D. Malo. 2006. Sequencing, expression, and functional analyses support the candidacy of Ncf2 in susceptibility to Salmonella typhimurium infection in wild-derived mice. J. Immunol. 1766954-6961. [DOI] [PubMed] [Google Scholar]
- 32.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 951669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaw, M. H., T. Reimer, Y. G. Kim, and G. Nunez. 2008. NOD-like receptors (NLRs): bona fide intracellular microbial sensors. Curr. Opin. Immunol. 20377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephan, K., I. Smirnova, B. Jacque, and A. Poltorak. 2007. Genetic analysis of the innate immune responses in wild-derived inbred strains of mice. Eur. J. Immunol. 37212-223. [DOI] [PubMed] [Google Scholar]
- 35.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 633609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe, T., M. Chamaillard, Y. Ogura, L. Zhu, S. Qiu, J. Masumoto, P. Ghosh, A. Moran, M. M. Predergast, G. Tromp, C. J. Williams, N. Inohara, and G. Nunez. 2004. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 231587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilney, L. G., O. S. Harb, P. S. Connelly, C. G. Robinson, and C. R. Roy. 2001. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J. Cell Sci. 1144637-4650. [DOI] [PubMed] [Google Scholar]
- 38.Tschopp, J., F. Martinon, and K. Burns. 2003. NALPs: a novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 495-104. [DOI] [PubMed] [Google Scholar]
- 39.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279873-876. [DOI] [PubMed] [Google Scholar]
- 40.Wilmanski, J. M., T. Petnicki-Ocwieja, and K. S. Kobayashi. 2008. NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J. Leukoc. Biol. 8313-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright, E. K., S. A. Goodart, J. D. Growney, V. Hadinoto, M. G. Endrizzi, E. M. Long, K. Sadigh, A. L. Abney, I. Bernstein-Hanley, and W. F. Dietrich. 2003. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr. Biol. 1327-36. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto, Y., T. W. Klein, and H. Friedman. 1992. Genetic control of macrophage susceptibility to infection by Legionella pneumophila. FEMS Microbiol. Immunol. 4137-145. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto, Y., T. W. Klein, C. A. Newton, and H. Friedman. 1988. Interaction of Legionella pneumophila with peritoneal macrophages from various mouse strains. Adv. Exp. Med. Biol. 23989-98. [DOI] [PubMed] [Google Scholar]
- 44.Zamboni, D. S., K. S. Kobayashi, T. Kohlsdorf, Y. Ogura, E. M. Long, R. E. Vance, K. Kuida, S. Mariathasan, V. M. Dixit, R. A. Flavell, W. F. Dietrich, and C. R. Roy. 2006. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7318-325. [DOI] [PubMed] [Google Scholar]