Abstract
The herpes simplex virus protein UL25 attaches to the external vertices of herpes simplex virus type 1 capsids and is required for the stable packaging of viral DNA. To define regions of the protein important for viral replication and capsid attachment, the 580-amino-acid UL25 open reading frame was disrupted by transposon mutagenesis. The UL25 mutants were assayed for complementation of a UL25 deletion virus, and in vitro-synthesized protein was tested for binding to UL25-deficient capsids. Of the 11 mutants analyzed, 4 did not complement growth of the UL25 deletion mutant, and analysis of these and additional mutants in the capsid-binding assay demonstrated that UL25 amino acids 1 to 50 were sufficient for capsid binding. Several UL25 mutations were transferred into recombinant viruses to analyze the effect of the mutations on UL25 capsid binding and on DNA cleavage and packaging. Studies of these mutants demonstrated that amino acids 1 to 50 of UL25 are essential for its stable interaction with capsids and that the C terminus is essential for DNA packaging and the production of infectious virus through its interactions with other viral packaging or tegument proteins. Analysis of viral DNA cleavage demonstrated that in the absence of a functional UL25 protein, aberrant cleavage takes place at the unique short end of the viral genome, resulting in truncated viral genomes that are not retained in capsids. Based on these observations, we propose a model where UL25 is required for the formation of DNA-containing capsids by acting to stabilize capsids that contain full-length viral genomes.
DNA packaging is a critical step in the lytic replicative cycle of herpes simplex virus type 1 (HSV-1). During the packaging reaction, replicated DNA is resolved from branched concatemers to single-unit-length genomes that are inserted into preformed capsids (10, 31). Three types of capsids are found in the nuclei of cells infected with HSV-1: A (empty), B (intermediate), and C (full) (reviewed in reference 14). The three are distinguishable morphologically in electron micrographs, and they can be separated from each other preparatively by sucrose density gradient ultracentrifugation. They differ in the material present inside the capsid cavity. C capsids contain the virus DNA, and A and B capsids lack DNA. The B capsid cavity is filled primarily with VP22a, the cleaved form of the scaffolding protein, while in A capsids, the cavity lacks both DNA and protein. While B capsids may form after spontaneous maturation of the procapsid, empty A capsids result from abortive DNA packaging (39). Seven HSV-1 genes have been identified that are required for DNA encapsidation (1, 2, 21-23, 27, 30, 34). Null mutants of the UL6, UL15, UL17, UL28, UL32, and UL33 genes produce only B capsids, and these mutations block cleavage of viral genomes from the replicated concatemer. In contrast, disruption of the UL25 gene results in the accumulation in the nucleus of both A and B capsids in addition to unpackaged, genome-length DNA (23, 33). Thus, UL25 is not required for cleavage or insertion of DNA into the capsid but rather for maintenance of the viral genome after the packaging event.
UL25 is a capsid-associated protein, and it is present on all capsid forms in increasing amounts from procapsid to B, A, and C capsids and virions (24, 32). UL25 attaches to the exterior capsid vertices in a proposed heterodimer with UL17 (35, 38). UL25 has been shown to interact with the triplex protein UL38 (VP19c) and the major capsid protein UL19 (VP5); these observations have been corroborated by cryoelectron microscopy and capsid reconstruction that predicts UL25 contacts triplexes and hexons that surround the penton (26, 38). It has been suggested that conformational changes of the capsid proteins induced by DNA packaging expose UL25 binding sites at the capsid surface (38). Much like the accessory proteins of the double-stranded DNA (dsDNA) bacteriophages, accumulation of UL25 protein may reinforce the capsid to prevent loss of DNA (23, 24). Finally, two recent studies have shown that UL25 is required for the attachment of the tegument to the capsid through its interaction with the large tegument protein UL36 (8) and that UL25 is required at a very early stage in infection, in the uncoating of the viral genome (28).
The UL25 gene product is a 580-amino-acid, 62-kDa protein that contains a flexible N-terminal tail and a more rigid core of packed alpha helices (5). In this study, we completed a mutational analysis of UL25 to define regions of UL25 protein important for viral replication and capsid attachment. We identified a UL25 capsid-binding domain that is located in the N terminus and is required for binding of UL25 to capsids and for successful DNA packaging in vivo. In addition, we isolated UL25 mutants that bind capsids in vivo without promoting the formation of infectious particles. These mutations may span regions of UL25 protein that are required for capsid maturation from the nucleus or other downstream events in virion assembly. These results indicate that UL25 capsid attachment is required but not sufficient for completion of the DNA packaging reaction. Finally, as previously proposed by Stow (33), the UL25 protein appears to play an important role during the later stages of DNA packaging prior to release of capsids into the cytoplasm. Here, we show that this late defect is due to the premature cleavage of the viral genome as it is being packaged, resulting in truncated genomes that failed to be retained in capsids.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
African green monkey kidney cells (Vero; American Type Culture Collection, Rockville, MD) and UL25-transformed 8-1 cells were propagated as previously described (23). HSV wild-type KOS and UL25-null virus KUL25NS have been previously described (23). The rabbit polyclonal antibody NC1 (7) was used in Western blots to detect the HSV-1 major capsid protein, UL19. Baculovirus-expressed UL25 protein (23) was purified and used to prepare a UL25 monoclonal antibody, 25E10, that was provided by W. Newcomb, University of Virginia (unpublished data).
Transposon mutagenesis.
The transposition reaction was done following the GPS-LS protocol (New England Biolabs). The GPS-LS system is an in vitro method for random insertion of a 1.7-kbp transposon (Tn7-based transprimer) that encodes a kanamycin resistance gene. The mutagenesis is accomplished by introduction of a transposon that contains the 8-bp PmeI site; the majority of the transposon is then removed by restriction digestion with PmeI. Religation results in a 15-bp insertion, which retains the unique PmeI site. Plasmid pAPV-UL25 contains the UL25 gene expressed from the HSV-1 ICP6 promoter and was used as the template for the transposition (23). Following transposition, the plasmid was transformed into bacteria and plated on selective agar containing kanamycin and ampicillin. DNA extracted from individual colonies was analyzed using a restriction enzyme (PmeI) to map the site of the transposition. Plasmids with insertions that mapped within the UL25 open reading frame (ORF) were digested with PmeI to remove transposon sequences, religated, and transformed into bacteria. The mutants were sequenced (University of Pittsburgh DNA sequencing core laboratory) to confirm the positions and residues inserted as a result of the transposition.
Construction of UL25 mutants.
UL25-expressing plasmids were made by PCR using pAPV-UL25 as a template followed by Topo cloning of the gel-purified PCR products into pcDNA3.1/V5/His-TOPO (Invitrogen) such that the UL25 gene was under the transcriptional control of the cytomegalovirus (CMV) promoter. PCR primers contained BamHI and EcoRI restriction sites that were used for further subcloning. UL25 plasmids pFH274, pFH284, pFH368, and pFH379 contained the coding sequences for UL25 amino acids 37 to 580, 1 to 580, 1 to 50, and 51 to 580, respectively. The following primers were used to amplify UL25: pFH274 (5′-GGATCCATGTCGCCCGTCTTTAACC-3′ and 5′-GGATCCCTAAACCGCCGACAGGTAC-3′), pFH284 (5′-GAATTCGGATCCATGGACCCGTACTGCCCATT-3′ and 5′-GGATCCAACCGCCGACAGGTACTGTGGAATG-3′), pFH368 (5′-GGATCCGAATTCATGGACCCGTACTGCCCATT-3′ and 5′-GGATCCCTCCGCCGCCGTCTCCCG-3′), and pFH379 (5′-GGATCCGAATTCATGCAGGTGGTCGTCCTACAGGCCCAGCGCACA-3′ and 5′-GGATCCAACCGCCGACAGGTACTGTGGAATG-3′).
The UL25 Topo clones were digested with BamHI, and UL25 sequences were then ligated into the BamHI site of plasmid pFH118. pFH118 expresses proteins (using the CMV promoter) with a C-terminal tandem affinity purification (TAP) tag and was constructed from plasmid pZOME-1-C (obtained from EUROSCARF, Institute of Microbiology, University of Frankfurt, Germany) (29). pZOME-1-C was digested with BamHI-EcoRI, and a 540-bp fragment containing the TAP ORF was cloned into pcDNA3 at the BamHI and EcoRI sites to generate pFH118. Plasmids pFH290, pFH373, and pFH392 expressed UL25-TAP fusion proteins 1-580-TAP, 1-50-TAP, and 51-580-TAP, respectively.
In vitro binding of UL25 to capsids.
Binding was performed with capsids lacking UL25 that were prepared from Vero cells infected as described below with the UL25-null mutant virus, vΔUL25. Synthesis of [35S]methionine-labeled UL25 was carried out in a rabbit reticulocyte lysate using a coupled transcription and translation system (TNT T7Quick; Promega) according to the manufacturer's instructions using pGEM-4z, pcDNA3.1/V5-HIS-TOPO, or pFH118 vectors expressing wild-type or mutant UL25 genes. The [35S]methionine-labeled UL25 (45 μl of the 50-μl in vitro translation reaction) was mixed with 50 μl of pooled A and B capsids plus 400 μl of phosphate-buffered saline (PBS), and the mixture was incubated for 90 min at room temperature with constant rotation. Capsids were isolated from the reaction mixture by centrifugation on a gradient of 20% to 50% sucrose in TNE (500 mM NaCl, 10 mM Tris, 1 mM EDTA [pH 7.5]) on an SW41 rotor at 24,000 rpm for 1 h at 4°C. After centrifugation, capsids were harvested from the gradient, precipitated by addition of an equal volume of 16% trichloroacetic acid, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were stained with Coomassie blue and dried onto Whatman 3M filter paper prior to autoradiography.
Construction of recombinant viruses.
Production and characterization of a bacterial artificial chromosome (BAC) containing the entire HSV-1 (KOS-37) genome was described previously (13). The KOS BAC clone (bFH439) containing a deletion that removed the entire UL25 ORF was produced with a counterselection BAC modification kit (Genebridges). Briefly, the Red/ET expression plasmid pSC101-BAD-gbaA (Genebridges) was transformed into EPI300 bacteria containing the wild-type KOS-37 BAC. After induction of Red/ET expression, cells were transformed with a PCR product consisting of the rpsL-neo antibiotic selection cassette flanked by sequences homologous to 50 bp immediately upstream and downstream of the UL25 ORF. Recombinant clones were isolated on selective agar medium, and deletion of UL25 from BAC DNA was confirmed by PCR with primers external to the recombination locus. To create specific UL25 mutations, the KOS BAC clone was transferred to GS1783 bacteria (gift from G. Smith) and mutagenesis was performed using the two-step bacteriophage λ Red-mediated homologous recombination system described by Tischer et al. (36). PCR was used to produce a construct that contained the Kanr gene (alphAI from plasmid p-EP-Kan-S) flanked by UL25 homologous sequence repeats containing the desired mutation or deletion. Integration of this construct into the BAC was followed by a second recombination that removed alphaAI and left the seamless mutation within the UL25 gene. The GFP coding sequence used for insertion within the UL25 ORF was amplified from plasmid peGFP-in (36). Resolution of the integrated PCR product resulted in insertion of the GFP gene between UL25 codons 50 and 51. The list of primers for production of PCR amplicons that were used for isolation of the UL25 deletion and insertion mutants can be found in Table 1. To recover mutant viruses, BAC DNA (5 μg) was transfected into 8-1 cells with Lipofectamine. Recombinant viruses harvested from transfected cell lysates were plaque purified on 8-1 or Vero cells.
TABLE 1.
Primers used for generation of recombinant HSV-BACs
| BAC | UL25 mutation | Primer sequencea |
|---|---|---|
| bFH439 | Deletion replaced | 5′-gacaacgaccgcagttctcgtgtgttattttcgctctccgcctctcgcag[agatct]GGCCTGGTGATGATGGCGGGATCG-3′ |
| with rpsL-neo | 5′-ggctcttgttttttctccctaatgccccctcccccctcgcccaccaccca[agatct]TCAGAAGAACTCGTCAAGAAGGCG-3′ | |
| bFH407 | 143i | 5′-gccgtcgcggagatggaggtccagatcgtgcgcaactgtttaaacagcaacgacccgccgTAGGGATAACAGGGTAATCGATTT-3′ |
| 5′-ggaggttggtgtcgtatcgtagcggcgggtcgttgctgtttaaacagttgcgcacgatGCCAGTGTTACAACCAATTAACC-3′ | ||
| bFH416 | 212s | 5′-actttcgggacggccggatgtccaagaccttcatgactgtttaaacaatgacggcgctggtTAGGGATAACAGGGTAATCGATTT-3′ |
| 5′-gccgcacgactgcagggacaggaccagcgccgtcattgtttaaacagtcatgaaggtctGCCAGTGTTACAACCAATTAACC-3′ | ||
| bFH418 | 560s | 5′-gctccaacaccaagcgtttctccgcgttcaacgttagtgtttaaacagttagcagcgactaTAGGGATAACAGGGTAATCGATTT-3′ |
| 5′-gacataaaaagtacaacatgtcgtagtcgctgctaactgtttaaacactaacgttgaacgGCCAGTGTTACAACCAATTAACC-3′ | ||
| bFH421 | Δ1-50 | 5′-accgcagttctcgtgtgttattttcgctctccgcctctcgatgcaggtggtcgtcctgcagTAGGGATAACAGGGTAATCGATTT-3′ |
| 5′-cggcagccgctgtgcgctgggcctgcaggacgaccacctgcatcgagaggcggagagcgaGCCAGTGTTACAACCAATTAACC-3′ | ||
| bFH422 | GFP insertion | 5′-gacttttggatgtcgcccgtctttaacctcccccgggagacggcggcggagATGGTGAGCAAGGGCGAGGA-3′ |
| 5′-ctccagggcagcggcagccgctgtgcgctgggcctgcaggacgaccacctgCTTGTACAGCTCGTCCATGCCG-3′ | ||
| bFH423 | 155i | 5′-cgacccgccgctacgatacgacaccaacctccccgtggtgtttaaacacgtggatctgcTAGGGATAACAGGGTAATCGATTT-3′ |
| 5′-ccgcggcccgcgtacaccatatgtagcagatccacgtgtttaaacaccacggggaggttgGCCAGTGTTACAACCAATTAACC-3′ |
[agatct], BglII restriction site. Mutations are underlined. Uppercase letters represent the template sequence (pEP-Kan-S and peGFP-in). Lowercase letters represent HSV UL25 gene sequences.
Southern blots.
A T-175 flask of 8-1 or Vero cells was infected with virus at a multiplicity of infection (MOI) of 5 PFU per cell. At 18 h postinfection, the medium was removed and the cells were washed in 1× PBS, scraped off the plate, and pelleted. The cells were lysed and viral DNA was prepared as previously described (15). The final DNA was digested with BamHI and PmeI to confirm the mutations and BamHI or HindIII to assess cleavage of viral DNA. DNA was separated by agarose gel electrophoreses, transferred to a nylon membrane, and hybridized as previously described (15). Southern blots were scanned with Storm 840 PhosphorImager, and specific bands were quantified with ImageQuant software.
Immunoblotting.
Protein extracts were separated on a 5 to 12% SDS-polyacrylamide gel, and proteins were transferred to nitrocellulose. The nitrocellulose was washed twice in Tris-buffered saline (TBS) and blocked overnight in TBS-T (20 mM Tris, 0.5 M NaCl [pH 7.5] plus 0.5% Tween 20) supplemented with 10% nonfat dry milk. UL25 monoclonal antibody, 25E10, and the VP5 rabbit polyclonal antibody, NC1, were diluted to 1:5,000 in blocking buffer. The diluted antibodies were reacted with the blocked nitrocellulose for 2 h at room temperature, washed, and reacted with horseradish peroxidase-conjugated anti-goat or anti-mouse immunoglobulin G (IgG) diluted at 1:5,000 in TBS-T plus 10% nonfat dry milk. The bound Igs were revealed by enhanced chemiluminescence (Pierce ECL kit).
Complementation assay.
Vero cells (1.0 × 105 cells/well in a 24-well plate) were transfected with 0.5 μg of plasmid DNA and 1 μl of Lipofectamine 2000 (Invitrogen) diluted in 100 μl of serum-free medium. The DNA-lipid complexes were allowed to form at room temperature for 20 min, added to the cells, and incubated overnight at 37°C. The next day, the cells were infected with UL25-null virus (vΔUL25) at an MOI of 5 PFU/cell. After 90 min at 37°C, the medium was removed and unabsorbed virus was inactivated by washing the cells three times with citrate buffer (pH 3). The cultures were incubated for an additional 18 h, after which the cells were scraped from the plate and rinsed with PBS. A 10% aliquot of total cells was frozen and used to determine virus titers on 8-1 cells and Vero cells. The remainder of the cell pellet was resuspended in 2× PAGE loading buffer (Invitrogen) and used for Western blot analysis.
Capsid purification.
Vero cells (1.5 × 108) were infected overnight (18 h at 37°C) at an MOI of 5 PFU/cell. Infected cells were harvested, rinsed with PBS, and resuspended in 20 mM Tris (pH 7.5) plus protease inhibitors (Roche), adjusted to 1% Triton X-100, and incubated for 30 min on ice. The resulting nuclei were harvested by low-speed centrifugation, resuspended in 10 ml TNE (500 mM NaCl, 10 mM Tris, 1 mM EDTA [pH 7.5]), and then sonicated to lyse the nuclei. The nuclear lysate was adjusted to 20 mM MgCl2 and incubated with DNase I (100 μg/ml) at room temperature for 30 min. The lysate was then cleared by low-speed centrifugation, and the resulting supernatant was layered on top of a 10-ml cushion of 35% sucrose (SW28 rotor; 23,000 rpm for 1 h). The pellets were suspended in 3 ml TNE and adjusted to 1 mM dithiothreitol, and capsids were separated by centrifugation on 20 to 50% sucrose (in TNE) gradients (SW41 rotor at 24,000 rpm for 1 h). The positions of A, B, or C capsids were observed as light-scattering bands, with A capsids being found highest (least dense) on the gradient and C capsids found lowest (dense fractions) on the gradients. The different capsid fractions can also be identified based on the presence or absence of the scaffold protein, VP22a, since only B capsids contain the scaffold protein. The fractions were collected using a Beckman fraction recovery system (Beckman catalog no. 34890). The apparatus has a mechanism to puncture the bottoms of the tubes, and fractions are collected from the bottom to top. Fractions (0.75 ml) were collected, and protein was precipitated by adding an equal volume of 16% trichloroacetic acid. Pellets were resuspended in 35 μl of 2× PAGE loading buffer (Invitrogen) supplemented with 0.4 M Tris-base. Gradient fractions were run on 5 to 12% SDS-PAGE, and the gels were either stained with Imperial blue (Pierce) to visualize capsid proteins or analyzed by immunoblotting with the UL25 (25E10) antibody.
RESULTS
Transposon mutagenesis and functional assessment of UL25 mutants.
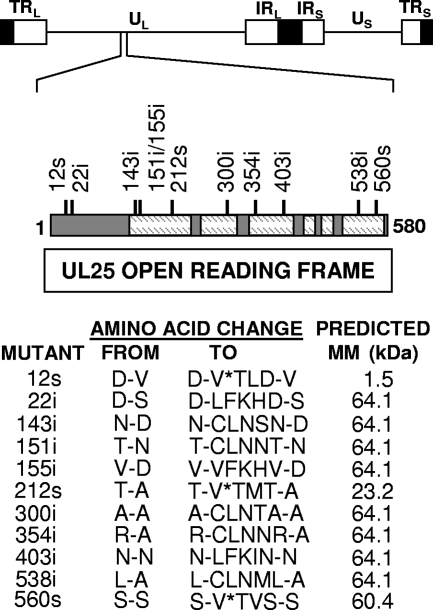
In order to identify regions of UL25 protein that are important for viral replication and for attachment to capsids, the UL25 ORF was randomly disrupted by linker scanning mutagenesis. A plasmid vector (pAPV-UL25) containing the UL25 gene was subjected to Tn7 transposon-mediated insertion of a 15-bp fragment that contained a PmeI restriction site. Eleven clones with unique DNA insertions in the coding region of the UL25 gene were mapped by restriction digestion and DNA sequencing. The resulting amino acid insertions into the UL25 protein are shown in Fig. 1. The mutations were scattered across the UL25 ORF with a clustering of three insertions between amino acids 143 and 155. Three of these mutants contained in-frame stop codons after amino acids 12, 212, and 560.
FIG. 1.
(Top) Physical map of the HSV-1 genome showing the location of the UL25 gene. UL and US refer to the long and short unique region sequences, respectively, and TRL and TRS and IRL and IRS refer to the terminal and internal repeat sequences, respectively. (Middle) Schematic representation of the 580-amino-acid UL25 protein. Dashed regions are the folded protein core as determined by characterization of the UL25 crystal structure (5). Numbers above the line indicate the amino acid location where 15 bp were inserted into the UL25 ORF, resulting in either the insertion (i) of five amino acids or an in-frame stop codon (s). (Bottom) The locations of the insertions and the predicted amino acid changes are shown along with the predicted molecular mass (MM) for each mutant. *, stop codon.
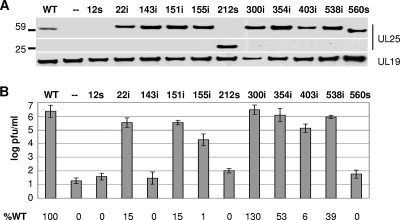
The ability of the UL25 transposon mutants to support virus replication was tested using a genetic complementation assay. Vero cells were transfected with wild-type and mutant plasmids, and the next day the cells were infected with the UL25-null virus, vΔUL25. Following another day of incubation, the cell lysates were harvested, tested for UL25 protein expression by Western blot analysis, and assayed for virus progeny yield by plating the lysates on UL25-complementing cells (8-1 cells). With the exception of the 12s mutant, all of the plasmids produced proteins of the expected sizes (Fig. 2A). Eight of the transposon mutants expressed a UL25 protein that was similar in size to the wild-type UL25 protein. For these mutants, a size difference would not be apparent since only five amino acids were added to the mutant UL25 proteins. The two nonsense mutants 212s and 560s produced proteins of 23 and 60 kDa, respectively. No immunoreactive proteins were observed for the 12s mutant, but this plasmid was predicted to make only a small peptide that would probably not be recognized by the UL25 antiserum. Using the HSV major capsid protein (UL19) as a loading control, all of the mutants except the 12s mutant were found to express similar amounts of UL25 (Fig. 2A). The 12s mutant contains a stop codon that would result in a truncated UL25 protein of only 12 amino acids that would not be detected on the gel. The ability of the UL25 mutants to complement growth of the UL25-null mutant was determined by titrating the cell lysates on 8-1 cells. Figure 2B shows that, with the exception of the three truncation mutants (12s, 212s, and 560s) and the 143i mutant, the remaining mutants were able to complement the UL25-null virus, although the 155i and 403i mutants complemented at less than 10% of the wild-type UL25 protein.
FIG. 2.
Complementation assays for the UL25 transposon mutants. The UL25 transposon mutants were examined for their ability to complement the growth of the UL25-null virus. (A) Western blots for UL25 protein and the major capsid protein, UL19. (B) The progeny virus was titrated on the UL25-complementing cell line, 8-1, and the results shown are averages of at least six experiments. Error bars represent the standard deviation. The data listed at the bottom are the percentages of complementation relative to the wild-type (WT) UL25 protein.
The N terminus of UL25 is required for capsid attachment in vitro.
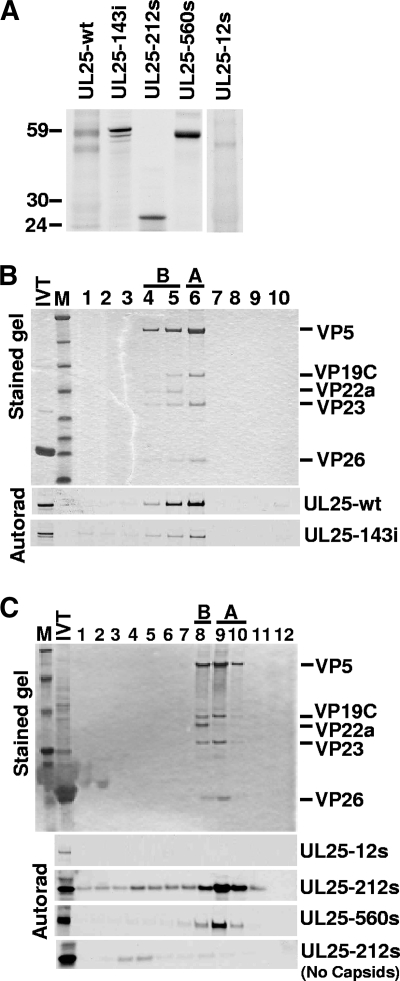
Previously we showed that [35S]methionine-labeled UL25 synthesized in a coupled in vitro transcription-translation system will attach to the surface of UL25-deficient capsids during in vitro incubation (24). To determine if mutations that eliminated complementation also affected UL25 capsid attachment, the 12s, 143i, 212s, and 560s mutants were tested for in vitro capsid binding. In vitro translation of plasmids containing the 143i, 212s, and 560s mutants produced proteins corresponding to their predicted molecular weights (Fig. 3A). Unexpectedly, in vitro translation of the 12s mutant produced a nearly wild-type-size protein (Fig. 3A). Capsid binding was determined by adding an aliquot of the [35S]methionine-labeled UL25 to pooled A and B capsids derived from the nuclei of cells infected with the UL25-null mutant, vΔUL25 (Table 2). After incubation to allow UL25 attachment, the capsids were separated from other reaction components by centrifugation on a sucrose gradient, and gradient fractions were analyzed by SDS-PAGE followed by autoradiography. Capsid attachment was indicated by comigration of UL25 protein with capsids in the sucrose gradient (Fig. 3B and C). The 143i, 212s, and 560s proteins attached to capsids. Although the 212s protein was found to be primarily associated with the fractions containing A and B capsids, significant levels of the protein were found in fractions denser than capsids. This was probably the result of aggregation of the truncated UL25 protein, as evidenced by the observation that this protein is found in the same dense fractions when the in vitro-translated protein was run on the gradient in the absence of A and B capsids (Fig. 3C). In contrast to the 143i, 212s, and 560s mutants, the 12s translation product did not attach to capsids, although we cannot rule out that there may be some low-level binding since the amount of radiolabeled 12s protein was reduced compared to the amounts of the other UL25 proteins. Examination of the UL25 coding sequence revealed that after the first AUG, the next in-frame AUG start codon was located ∼120 bp downstream. This start site corresponded to UL25 amino acid M37. To determine if the 12s mutant was initiating at this downstream start site, the codons for amino acids 1 to 36 were deleted from the UL25 expression plasmid (Fig. 4A). The resulting in vitro-synthesized protein, containing UL25 amino acids 37 to 580, was found to be identical in size to the 12s mutant in vitro translation product (Fig. 4B). Similar to the result with the 12s protein, the UL25 N-terminal truncation (37 to 580) mutant did not bind capsids when tested in the in vitro capsid-binding assay (Fig. 4C). In addition, this mutant did not complement the UL25-null virus (data not shown). These results demonstrated that the sequences within the N terminus of the UL25 protein are required for capsid binding.
FIG. 3.
Capsid binding by UL25 transposon mutants. (A) Autoradiograph of [35S]Met-labeled UL25 protein after SDS-PAGE. The sizes of the 143i, 212s, and 560s proteins were consistent with predicted molecular mass for each protein, but the 12s mutant unexpectedly produced a nearly wild-type(wt)-size UL25 protein. (B and C) In vitro capsid-binding assay. In vitro-translated protein was incubated with pooled A and B capsids isolated from Vero cells infected with the UL25-null virus, vΔUL25, and capsids were then purified by sucrose gradient centrifugation. The sucrose gradients were fractionated and analyzed by SDS-PAGE followed by Coomassie staining (top panel, representative gel) and autoradiography (bottom panels). The fractions containing A and B capsids are indicated. The capsid proteins visible in the stained gel are listed at right. Capsid binding was demonstrated by comigration of the UL25 protein with A and B capsids. Aggregation of the truncated 212s protein may explain the presence of this protein in the same dense fractions as in the gradient lacking A and B capsids (bottom of panel C). IVT, in vitro transcription-translation; M, molecular mass markers (from top, 194, 104, 60, 41, 27, 21, 16, and 7 kDa). Lane 1 is the bottom of the gradient and lane 10 (B) or 12 (C) is the top.
TABLE 2.
Virus growth assay results
| Virus strain | UL25 protein expressed (amino acid positions) | Virus growth (PFU/ml) in:
|
|
|---|---|---|---|
| Vero cells | 8-1 cells | ||
| KOS | Wild type (1-580) | 2 × 108 | 2 × 108 |
| KUL25NS | Null (1-107) | 3 × 104 | 2 × 108 |
| vΔUL25 | Null | <1 × 102 | 9 × 107 |
| v143i | 143i (1-580) | <1 × 102 | 3 × 107 |
| v212s | 212s (1-212) | <1 × 102 | 1 × 107 |
| v560s | 560s (1-560) | <1 × 102 | 2.7 × 107 |
| vΔ1-50 | Δ1-50 (51-580) | <1 × 102 | 3 × 108 |
| vUL25-GFP | UL25-GFP (1-50-GFP-51-580) | 4 × 107 | 4 × 107 |
| v155i | 155i (1-580) | 1 × 107 | 6 × 107 |
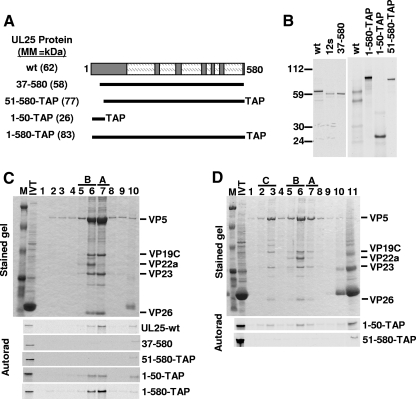
FIG. 4.
Capsid binding by UL25 N-terminal mutants. (A) UL25 N-terminal truncation and TAP fusion constructs are shown. The numbers listed on the left indicate the UL25 amino acids included in each construct. The TAP constructs contained the TAP tag fused to the C terminus of UL25. The predicted molecular mass (MM) of each protein is listed in parentheses. wt, wild type. (B) Autoradiograph of in vitro translation (IVT) products after SDS-PAGE. Molecular mass markers in kDa are shown at left. Each protein showed the appropriate molecular mass due to either addition of the TAP tag or deletion of the UL25 N-terminal amino acids. (C) In vitro capsid-binding assay. In vitro-translated protein was incubated with pooled A and B capsids isolated from Vero cells infected with the UL25-null virus, vΔUL25, and then purified by sucrose gradient centrifugation. A representative Coomassie-stained gel of the gradient fractions is shown (top panel). Autoradiographs of dried gels demonstrating the binding of 1-580-TAP or 1-50-TAP but not 51-580-TAP or the 37-580 to the two capsid types (bottom panel). (D) In vitro-translated protein was incubated with pooled wild-type KOS A, B, and C capsids and then purified by sucrose gradient centrifugation. A representative stained gel of the gradient fractions is shown (top panel). Autoradiographs of dried gels demonstrating the binding of 1-50-TAP but not 51-580-TAP to all three capsid types (bottom panel). The fractions containing A, B, and C capsids are indicated. The capsid proteins visible in the stained gel are listed to the right. IVT, in vitro transcription-translation; M, molecular mass markers (from top, 194, 109, 59, 30, 22, 13, and 6 kDa). Lane 1 is the bottom of the gradient, and lane 10 or 11 is the top.
To determine if the UL25 N terminus was sufficient for capsid binding, three plasmids were generated that expressed portions of UL25 protein fused at its C terminus to a TAP tag (Fig. 4A). The TAP tag consists of the IgG-binding peptide protein A, the cleavage site for the tobacco etch virus protease, and the calmodulin binding domain; the tag adds 178 amino acids (∼20 kDa) to the C terminus of the UL25 fusion protein (29). The three constructs 1-580-TAP, 51-580-TAP, and 1-50-TAP contained the full-length UL25 protein, UL25 amino acids 51 to 580, and UL25 amino acids 1 to 50 fused to the TAP tag, respectively (Fig. 4A). In vitro translation produced proteins corresponding to their predicted size, and each protein was tested for binding to UL25-deficient A and B capsids (Fig. 4A to C). Like the 12s and 37-580 mutants, the UL25 fusion protein 51-580-TAP did not attach to capsids. In contrast, the full-length 1-580-TAP and truncated 1-50-TAP fusion proteins were able to bind capsids. When incubated with A, B, and C capsids purified from KOS-infected cells, the 1-50-TAP but not the 51-580-TAP was found to bind all three capsid types by binding to unoccupied sites on the capsids and/or competitively displacing wild-type protein from capsids (Fig. 4D). These results demonstrated that UL25 amino acids 1 to 50 mediate capsid attachment in the in vitro capsid-binding assay.
Introduction of UL25 mutations into the virus genome.
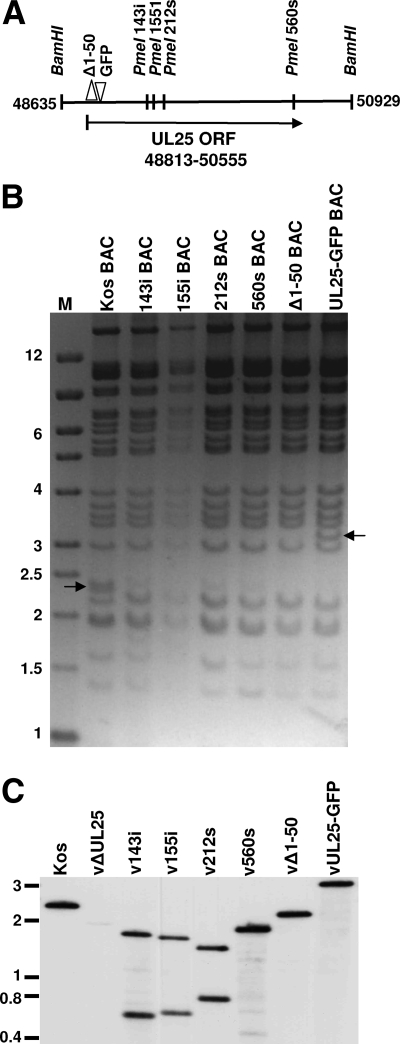
Several of the UL25 mutations were introduced into the viral genome through the genetic manipulation of an HSV-1 (KOS) genome maintained in a recombinant bacterial artificial chromosome (BAC). The genotypes of the viruses used in these studies are listed in Table 2. Virus vΔUL25 contains the rpsL-neo antibiotic resistance genes, which replace the entire UL25 ORF. Recombinant viruses v143i, v155i, v212s, and v560s contain the indicated UL25 linker insertion mutations (Fig. 5A). Recombinant virus vΔ1-50 contains a deletion that removed the coding sequences for UL25 amino acids 1 to 50, which were replaced with an ATG start codon. Recombinant virus vUL25-GFP contains DNA that encodes the GFP gene inserted in frame between codons 50 and 51 of the UL25 gene (Fig. 5A). The BACs used to generate each virus were confirmed by digesting them with BamHI and PmeI. The UL25 ORF is contained within the HSV-1 2,300-bp BamHI U fragment (Fig. 5A). Since there are no PmeI sites in the HSV-1 genome, the presence of the PmeI linker in the UL25 gene of the HSV BACs 143i, 155i, 212s, and 560s resulted in the loss of the 2,300-bp BamHI U fragment, which was shifted to a smaller fragment that comigrates with other HSV-1 BamHI fragments (Fig. 5B). The HSV BACs Δ1-50 and UL25-GFP contained the deletion of 150 bp and the insertion of 726 bp in UL25, respectively, which were detected as shifts in the BamHI U fragments (Fig. 5B).
FIG. 5.
Characterization of BAC DNA and HSV-infected cell DNA. (A) Schematic representation of the BamHI region located between nucleotides 48635 and 50929 of the HSV-1 genome that contains the UL25 ORF. The sites where unique PmeI restriction sites are found in the UL25 gene and the resulting BACs and viruses (143i BAC, 155i BAC, 212s BAC, and 560s BAC) are listed on top. Δ1-50 BAC contained a deletion that removes the coding sequences for UL25 amino acids 1 to 50 and inserts a new ATG start codon. UL25-GFP BAC contained an in-frame insertion of the (726-bp) GFP coding sequence after the codon for UL25 amino acid 50. (B) BAC DNA was digested with BamHI and PmeI and run on a 1.2% agarose gel. The wild-type 2.3-kbp BamHI fragment is present in the KOS BAC lane (arrow). This fragment was cleaved to smaller fragments by PmeI in 143i BAC, 155i BAC, 212s BAC, and 560s BAC. The UL25 fragments of Δ1-50 BAC and UL25-GFP BAC are shifted to 2.1 kbp (which comigrates with another viral band) and 3.2 kbp (arrow), respectively. M, DNA markers in kbp. (C) Southern blot of viral DNA. Viral DNA was purified from cells infected with the indicated viruses. Viral DNA (10 μg) was digested with BamHI and PmeI and run on a 1.2% agarose gel. DNA was blotted to a nylon membrane, and the blot was hybridized with the 32P-labeled UL25-containing BamHI U fragment. The wild-type BamHI fragment containing UL25 is visible at 2.3 kbp in the KOS lane and was cleaved to smaller fragments by PmeI in v143i, v155i, v212s, and v560s. The UL25 fragments of vΔ1-50 and vUL25-GFP are shifted to 2.1 and 3.2 kbp, respectively.
The BACs were transfected onto UL25-complementing 8-1 cells, and the recovered viruses were plaque purified on Vero or 8-1 cells. Each mutant was titrated on Vero and 8-1 cells to determine the effects of UL25 mutations on viral replication (Table 2). The vUL25-GFP mutant was able to replicate on Vero cells, while Vero cells were nonpermissive for the replication of vΔUL25, v143i, v212s, v560s, and vΔ1-50 (Table 2). The virus v155i displayed an intermediate phenotype with a sixfold reduction of growth on Vero cells as compared to the UL25-complementing cell line. To verify the presence of the mutations in the progeny virus, viral DNA was digested with BamHI and PmeI and subjected to Southern blot analysis with the 2,300-bp BamHI fragment (32P labeled) UL25 probe (Fig. 5C). As seen in the restriction enzyme digestion analyses of the individual BAC preparations, a prominent 2,300-bp fragment was detected in the KOS lane, which was reduced to two smaller fragments in v143i, v155i, v212s, and v560s and one smaller fragment in vΔ1-50. The 400-bp fragment for v560s was observed on longer exposure (not shown). The 2,300-bp fragment containing the UL25 ORF was not detected in the UL25 deletion virus vΔUL25. The insertion of the 726-bp GFP gene in the vUL25-GFP virus resulted in a larger BamHI fragment. These results demonstrated that that the UL25 mutants had the desired insertions or deletions.
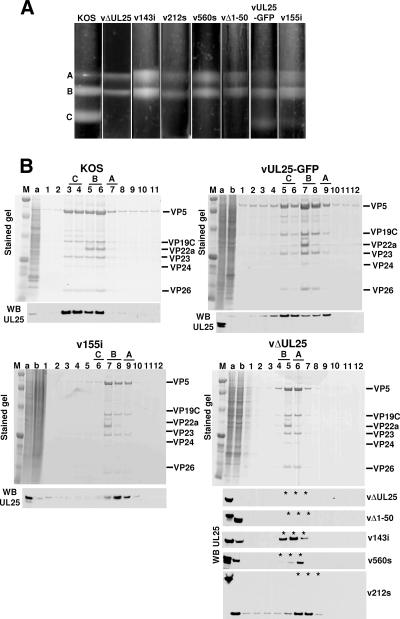
Association of UL25 with purified capsids.
Intranuclear capsids were purified from Vero cells infected with wild-type KOS or UL25 mutant viruses. The nuclear lysates were subjected to sucrose gradient centrifugation, and capsids were viewed as visible light-scattering bands (Fig. 6A). In cells infected with wild-type KOS or the two UL25 mutants vUL25-GFP and v155i, which replicate on Vero cells, all three capsid forms were observed. The KOS gradients contained nearly equal amounts of both B and C capsids, with only minor amounts of A capsids (Fig. 6A). In contrast, the gradients for the vUL25-GFP and v155i mutants contained less C capsids and more A and B capsids, and the C capsid band was barely detectable in the v155i gradient. The lysates of the UL25 mutants that did not replicate on Vero cells (vΔUL25, v143i, v212s, v560s, vΔ1-50, and KUL25NS) contained approximately equal numbers of A and B capsids but no DNA-containing C capsids (Fig. 6). The high proportion of A capsids found along with the absence of C capsids in all of these UL25 mutants is identical to what was previously described with the UL25-null mutant KUL25NS (23). To determine whether the UL25 protein was present in these capsids, the sucrose gradient fractions were analyzed by Western blot analysis to detect the UL25 protein (Fig. 6). The KOS gradient showed that the UL25 protein was found in the fractions that contained A, B, and C capsids. This result was replicated in capsids purified from cells infected with the vUL25-GFP virus. The 155i mutant was found to associate with A and B capsids, but since there were so few C capsids present on the gradient, there was little if any of this protein detected with these DNA-containing capsids. The UL25 linker insertion mutant 143i and the nonsense mutants 212s and 560s were found associated with A and B capsids isolated from cells infected with viruses expressing those proteins. The vΔ1-50 mutant expressed a truncated UL25 protein that was missing residues 1 to 50, and although this protein could be detected in the cell extract, it failed to bind either the A or B capsids. This result demonstrates the importance of the N terminus of the UL25 protein for capsid association of UL25. These data demonstrate that similar to the result of the in vitro capsid-binding assay (Fig. 3), the UL25 capsid-binding domain resides within the first 50 amino acids of the UL25 protein. Furthermore, the studies with the UL25 mutant viruses demonstrate that the UL25 capsid-binding domain is essential but not sufficient for production of infectious virus, as both virus progeny and C capsids were significantly reduced in noncomplementing cells infected with the 155i mutant (Table 2).
FIG. 6.
Analysis of capsid-bound UL25. Vero cells were infected at an MOI of 5 with KOS or the indicated UL25 mutants. Nuclear lysates were subjected to sucrose gradient sedimentation. (A) Capsids from Vero cells infected (MOI of 5) with KOS or the indicated UL25 mutant virus were harvested at 18 h postinfection, layered onto 20 to 50% sucrose gradients, and centrifuged at 24,000 rpm (SW41 rotor) for 1 h. The positions of A, B, and C capsid bands are indicated. (B) The protein composition for each gradient fraction was determined by SDS-PAGE (stained gel). Lane 1 is the bottom of the gradient, and lane 11 or 12 is the top. Gradient fractions that contained A, B, and C capsids are indicated. The positions of the capsid proteins (major capsid protein, VP5; triplex proteins, VP19C and VP23; scaffold protein, VP22a; smallest capsid protein, VP26) are indicated. (Bottom panels) Gradient fractions were analyzed by SDS-PAGE followed by immunoblotting for UL25 using the 25E10 antibody. The stained gel in the bottom right panel contains capsids isolated from UL25-null virus vΔUL25, which is representative of the mutants (vΔ1-50, v143i, v212s, and v560s) that fail to replicate on Vero cells. The UL25 Western blots (WB) for these mutants are shown below the stained gel with asterisks indicating the fractions that are equivalent to the capsid-containing fractions (lanes 5 to 7) of the vΔUL25-stained gel. Lane a, cell lysate from KOS-infected cells; lane b, cell lysate from UL25 mutant-infected cells. Molecular mass standards are visible in lanes M (from top, 194, 109, 59, 30, 22, 13, and 6 kDa).
Replication and cleavage of HSV-1 DNA.
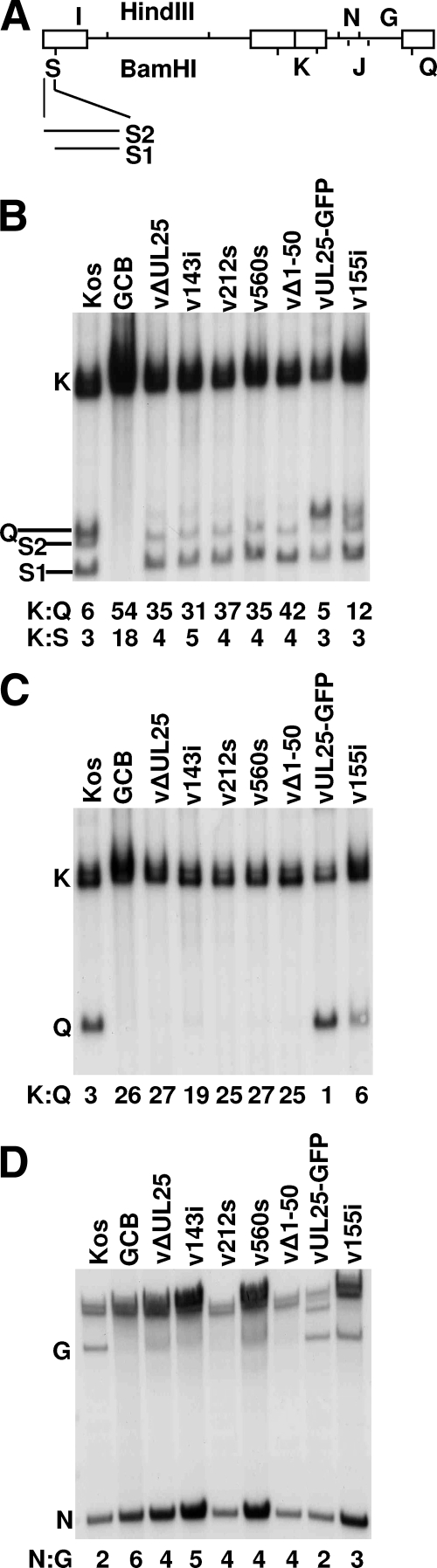
Previously, it has been shown that the UL25-null virus KUL25NS was able to cleave replicated viral DNA into unit-length genomes but failed to stably package the cleaved genomes (23, 33). To determine whether viruses bearing linker insertions or nonsense mutations were impaired in DNA cleavage and packaging, total DNA was isolated from Vero cells infected with wild-type and mutant viruses and subjected to Southern blot analysis. Viral DNA replication generates concatemers that are cleaved into unit-length molecules and packaged into virions. The cleaved viral DNA contains free chromosomal termini, and the presence of chromosomal ends can be monitored by Southern blot analysis of total infected cell DNA digested with BamHI and probed with the HSV-1 BamHI K fragment. Only cleaved viral DNA with free chromosomal ends will give rise to the terminal BamHI Q and S fragments, while concatemeric DNA gives rise to only the joint-spanning BamHI K fragment (Fig. 7A). The Q and S terminal fragments were detected in Vero cells infected with the wild type or with the two UL25 mutants (vUL25-GFP and v155i) that replicate on Vero cells. The S fragment represents the unique long (UL) terminus, and the Q fragment represents the unique short (US) terminus (Fig. 7A). The different sizes of the BamHI S and K fragments are due to the presence of one to multiple copies of the a sequences, while the Q fragment contains only a single copy of the a sequence. An example of a mutant that fails to cleave replicated viral DNA is the null mutant GCB, which does not express the product of the HSV UL28 gene (34). As shown in Fig. 7B, the terminal Q and S fragments were absent from DNA isolated from Vero cells infected with GCB. All of the UL25 mutants replicated viral DNA when grown on Vero cells, as assessed by the presence of the joint K fragments, but DNA isolated from Vero cells infected with replication-defective UL25 mutants lacked the terminal Q fragments (Fig. 7B). Since the Q and S fragments are similar in size, the blot was probed with a 32P-labeled fragment that hybridizes specifically to the US terminus. The probe detected the Q fragment in the digests for the three viruses that replicate on Vero cells but not in any of the viruses that do not; the junction fragment K was present in all samples (Fig. 7C). The amounts of radioactivity in the bands corresponding to the joint fragment, the long terminus, and the short terminus (BamHI fragments K, S, and Q, respectively) were measured, and the ratio of joint to each terminal fragment was calculated for each lane in Fig. 7B. In the case of the BamHI S fragments from the UL terminus, the measurement included both the S1 and S2 fragments. For wild-type HSV-1, the ratios of joint to end fragments were similar. However, for the UL25-null virus, vΔUL25, and the other UL25 mutants that do not replicate in Vero cells, the UL terminus was present at levels similar to those in the wild-type virus, but the US terminus was absent or significantly underrepresented compared to the joint fragment (Fig. 7B). The ratios of the end fragments for DNA isolated from cells infected with vUL25-GFP were similar to that of KOS, while there was a twofold reduction in the Us terminus for the 155i linker insertion mutant, v155i. This is consistent with the fact that replication of v155i is reduced (Table 2) and demonstrates a direct correlation of the defect with the cleavage of viral DNA, the production of stable C capsids, and the production of infectious virus.
FIG. 7.
Processing of virus DNA. (A) Schematic diagram of the HSV genome showing the locations of the HindIII G, I, and N fragments and the BamHI J, K, Q, and S fragments. The different sizes of the BamHI S fragment are due to the presence of one to multiple copies of the a sequence at the UL terminus. (B to D) Vero cells were infected with the indicated virus at an MOI of 5 PFU per cell. At 18 h postinfection, total infected cell DNA was isolated, digested with BamHI (B and C) or HindIII (D), and subjected to Southern blot analysis. The blots were probed with 32P-labeled BamHI K (B), BamHI Q (C), or BamHI J (D) fragments of the HSV-1 genome. The ratio of the BamHI joint K fragment to US-end Q or UL-end S fragments and the ratio of the internal HindIII N fragment to the US-end G fragment were determined by quantification of the hybridizing bands with phosphorimaging software.
The absence of US terminal fragments from the UL25 mutants was surprising, because previous studies from our laboratory demonstrated that genome-size (152 kbp) HSV DNA is present in cells infected with the UL25-null virus, KUL25NS. To further examine the US terminus generated by the UL25 mutants, total infected cell DNA was digested with HindIII and subsequently analyzed by Southern blot analysis using the HSV-1 BamHI J fragment as a probe (Fig. 7A). The probe spanned the HindIII restriction site that links the genome fragments containing the 13-kbp fragment from the US terminus (HindIII G fragment) and an internal 5-kbp fragment from the US region (HindIII N fragment).
The HindIII G fragment was readily detected in DNA isolated from Vero cells infected with KOS and the two UL25 mutants that replicate in Vero cells (vUL25-GFP and v155i), but this fragment was absent from DNA isolated from Vero cells infected with the UL28 mutant, GCB (Fig. 7D). Although the HindIII G fragment was present in the blots of DNA isolated from the five UL25 mutants (vΔUL25, v143i, v212s, v560s, and vΔ1-50) that fail to grow on Vero cells, the hybridizing band was diffuse, suggesting that the probe detected multiple US terminal fragments (Fig. 7D). The presence of multiple US terminal fragments would indicate that either the cleavage reaction is taking place at several sites to generate HindIII G fragments of different sizes or that the full-length genomic DNA is not stable and is being degraded. Because the BamHI Q fragment is absent from all of the UL25-defective mutants, the US terminus for these mutants appears to be located at least 3,000 bp 5′ to the wild-type US terminus. The ratio of HindIII G fragment to the internal HindIII N fragment was calculated for each virus and showed that the cleavage at the US end is reduced approximately twofold for the UL25 mutants. The two large hybridizing fragments that are present in all of the samples are viral DNA fragments that result from cleavage of the replicated DNA concatemers and consist of the joint-spanning fragments HindIII G plus HindIII B or, when the UL is inverted, HindIII G plus HindIII I (Fig. 7A).
DISCUSSION
The HSV-1 UL25 protein is one of seven viral proteins that are required for DNA cleavage and packaging. UL25 is found at or near each of the vertices of DNA-containing C capsids on the capsid surface. The capsid-bound UL25 appears to be a multifunctional protein required: (i) to stabilize capsids after DNA is packaged; (ii) to trigger nuclear egress of the capsid once a stable DNA-containing capsid is formed; (iii) to anchor VP1/2, the major tegument protein to capsids; and (iv) to uncoat the incoming viral genome early during infection (8, 23, 24, 28, 33, 38). Random mutagenesis of the HSV-1 UL25 gene was used to identify regions of UL25 protein important for virus replication and for UL25 capsid attachment. Although the five amino acid insertions disrupted core portions of the folded protein, most UL25 transposon mutants were able to complement the UL25 deletion virus. Four mutants were defective in the complementation. Insertion of stop codons after amino acids 212 and 560 resulted in the expression of nonfunctional truncated proteins that attached to capsids in vitro and when expressed from a recombinant virus. Similarly the 143i insertion mutant expressed a nonfunctional protein that failed to complement the UL25-null virus but retained the ability to bind capsids. The 12s mutant failed to complement and was predicted to encode a small UL25 peptide, but instead, in vitro translation of 12s produced a nearly wild-type-size protein that did not bind capsids. Examination of the UL25 DNA sequence indicated that the resulting protein was probably the result of translation initiation at an internal start codon (M37) in the UL25 gene. This result was confirmed by constructing a UL25 mutant in which the codons for amino acids 1 to 37 were deleted and replaced with an ATG codon. In vitro translation of this construct produced a protein the same size as the 12s mutant that also failed to bind capsids. Deletion of the UL25 N terminus to amino acid 50 eliminated in vitro capsid binding; conversely, the UL25-TAP fusion protein containing UL25 amino acids 1 to 50 was able to bind capsids in vitro. A recombinant virus expressing the UL25 51-580 protein was replication defective and produced capsids lacking the virus-expressed mutant protein. The fact that insertion of GFP between UL25 amino acids 50 and 51 did not affect virus replication suggests that the capsid binding domain resides solely within amino acids 1 to 50. This conclusion is supported by the capsid-binding studies where 1-50-TAP was shown to bind capsids (Fig. 4C). This result indicated that separation of amino acids 1 to 50 from the rest of the UL25 N terminus did not preclude capsid attachment or virus replication. Taken together, these data indicate that amino acids 1 to 50 are necessary and sufficient for UL25 capsid binding and therefore comprise the UL25 capsid-binding domain. However, binding of UL25 to the capsid through the N-terminal capsid-binding domain is not sufficient for virus production. The two truncation mutants v212s and v560s both express proteins that associate with capsids, but these mutants do not produce infectious virus, indicating that the final 19 amino acids of UL25 are also essential for function.
Mutational analysis of UL25 structure.
The crystal structure of UL25 (amino acids 134 to 580) has been determined (5). The UL25 protein consists of numerous flexible loops extending from a rigid core composed of several tightly packed alpha helices. We expected that mutations disrupting any of the alpha helices located in the core would alter the overall structure of the protein and hence inhibit its function. However, most of the mutants isolated in this study contained insertions in the UL25 core that did not reduce function of the plasmid-expressed mutant protein. For example, the 354i and 538i mutations were both located in internal alpha helices, but neither of these mutations caused significant reduction in complementation of the UL25-null virus. The 143i, 151i, and 155i insertion mutations occur within a region where the predicted amino acid sequence is strongly conserved within homologues of the UL25 gene of other herpesviruses. In the folded protein, this region is part of a loop that bridges the unstructured N terminus (residues 1 to 45) with the highly structured C terminus (residues 133 to 580); there are two small antiparallel beta sheets located between amino acids 133 and 155 (5). The 143i insertion maps to the turn between the two beta sheets, and the 151i insertion occurs shortly after the end of the second beta sheet. The 155i insertion is located within the first alpha helix found in the core structure. Interestingly, the 143i and 155i insertions altered virus replication, but the 151i insertion did not. Our results suggest that this region does not contribute to capsid binding but is important for UL25 function, possibly as a bridge from the flexible capsid-binding region (residues 1 to 50) of UL25 to the more structured C terminus. Other structural features of UL25 protein include electropositive and electronegative faces and clusters of conserved surface residues (5). The 151i and 155i mutations are located in the electropositive face of UL25, while the other mutations occur along the periphery or in the interior of the folded protein. The 151i and 155i mutations also fall within one of the conserved surface clusters identified by Bowman et al. (5).
HSV-1 DNA packaging.
In its basic features, the pathway for assembly of the HSV-1 capsid resembles that observed in dsDNA bacteriophages (3, 11, 19). The process of DNA encapsidation in bacteriophages begins when the terminase makes a double-strand cut in the replicated concatemer DNA and binds to the end of the DNA. The terminase-DNA complex then docks onto the procapsid at the portal vertex. DNA is then driven into the capsid by the terminase complex until it encounters a second cut site in the concatemer DNA. The second cut may occur at a second packaging signal or after a headful of DNA has been injected. As DNA is entering, the procapsid is transformed into its mature, icosahedral morphology. After the last DNA end has entered the capsid, the portal is closed, and the capsid is stabilized by addition of head completion proteins.
HSV DNA translocation into the capsid is mediated by the terminase complex comprised of the HSV UL15, UL28, and UL33 proteins (18, 40, 41). The cis-acting HSV-1 packaging sequences, designated pac1 and pac2, are found near the genomic ends of viral DNA. At the UL end of the genome, the pac2 site defines packaging directionality by mediating initiation, and pac1 serves to terminate packaging at the US end after a unit-length genome has entered the capsid (6). Terminase cleaves viral DNA at the pac2 site and then docks the free end of DNA at the capsid portal vertex. Packaging proceeds from the UL end until a full-length genome has entered the capsid; DNA cleavage occurs at a pac1 site at the US end. The packaged genome is then sealed within the capsid. Similar to the dsDNA bacteriophages, DNA cleavage is suppressed until a full genome (headful mechanism) has entered the capsid because otherwise shorter packaged genomes would be formed from cleavage at the packaging sequences within the HSV genome's internal repeat sequences. The addition of UL25 to the capsid at or near the end of the packaging process suggests that it may function similarly to lambda phage head completion proteins, gpD, or the Soc and Hoc proteins of phage T4; these proteins bind to the phage capsid surface after DNA has entered and are thought to reinforce the capsid structure (12, 16, 17, 42). Although most of our UL25 mutant proteins were able to attach to capsids, the failure of recombinant viruses to replicate on Vero cells was coincident with premature cleavage of the US end of the viral genome, as previously described for the UL25-null mutant KUL25NS (28, 33). In CMV, the halogenated benzimidazoles BDCRB (2-bromo-5,6-dichloro-1-β-d-riborfuranosyl benzamidazole riboside) and TCRB (2,5,6-trichloro-1-β-d-riborfuranosyl benzimidazole riboside) induce premature cleavage events during DNA packaging such that the site of cleavage is located at a similar distance from the end of the genome as was observed with the UL25 mutants (25). These compounds are not active against HSV-1, but resistance mutations to BDCRB and TCRB map to the human CMV terminase genes UL89 and UL56, which are the homologues of HSV-1 UL15 and UL28, respectively (20, 39). While the drugs’ mechanism of action is not known, they may relax the cleavage site sequence specificity of the terminase without relieving the headful requirement. By extension, the absence of functional UL25 on the capsid surface may relax the cleavage site specificity and allow aberrant cleavage by the terminase.
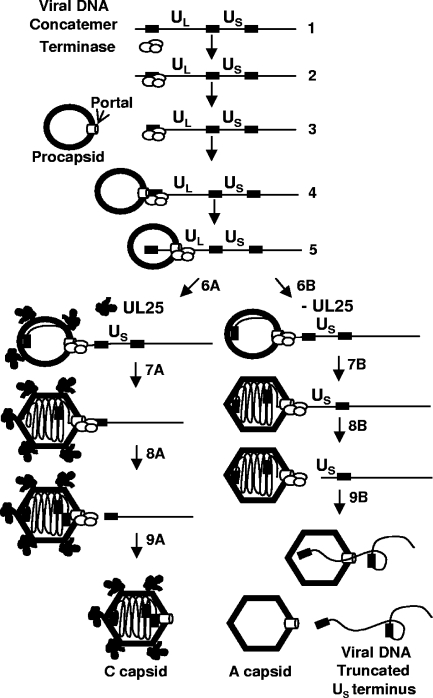
Role of UL25 in DNA packaging.
In conclusion, we propose to integrate our current studies of UL25 into the HSV DNA encapsidation model (Fig. 8). DNA packaging is an energy-dependent process that requires conformational changes in the capsid proteins and in the viral DNA itself (4, 9, 37). Presumably, terminase supplies the force required to compact DNA within the capsid. UL25 association with the capsid may stabilize this transition, such that in the absence of functional UL25, the equilibrium between the internal pressure of packaged DNA and the force exerted by the terminase is breached. When the terminase can no longer propel DNA into the nascent capsid, it stalls on the DNA and cleaves to abort packaging, forming the truncated genomes observed in replication-deficient UL25 mutants. The UL25 capsid-binding domain is responsible for the binding of UL25 to the capsid surface, but other portions of the protein are essential for completion of DNA packaging. These essential functions could include a direct role in the cleavage reaction, either by UL25 interaction with the terminase and portal complexes or by UL25 binding to the pac1 site.
FIG. 8.
Model for the role of UL25 in DNA packaging. Proposed model for the role of UL25 in HSV DNA encapsidation. (Step 1) DNA packaging initiates on concatemers when the terminase complex consisting of UL15, UL28, and UL33 binds (step 2) to packaging sequences and cleaves (step 3) the concatemer to generate a UL end with the terminase still bound. The terminase-DNA complex then docks at the portal vertex on the procapsid (step 4), and the terminase initiates DNA packaging (step 5). As the procapsid is filled (step 6A) with DNA, it angularizes and in the process, UL25 binding sites are exposed at the capsid vertexes. DNA cleavage is suppressed by a headful mechanism (step 7A) until a full-length genome has entered. Cleavage occurs at a packaging site located at the US end of the genome (step 8A). More UL25 binds to the capsid, and the newly packaged genome is sealed within the C capsid (step 9A). In the absence of a functional UL25 protein, the packaging reaction proceeds (step 6B) with the capsid filling and expanding until close to a genome length has entered (step 7B). Without UL25 to stabilize the capsid against the pressure that the packaged genome generates, premature cleavage (step 8B) occurs just prior to the entry of the US-end repeat, resulting in genomes with truncated US termini. In the absence of UL25, the capsid is not stable. The DNA is released (step 9B), which generates an empty A capsid and a free viral genome that is truncated at the US end.
Acknowledgments
We thank Klaus Osterrieder and David Leib for BAC reagents, Greg Smith for the GS1783 bacterial strain, Jamie Huffman for technical assistance, and Jay Brown for critically reading the manuscript.
This work was supported by Public Health Service grant AI060836 from the National Institutes of Health.
Footnotes
Published ahead of print on 22 October 2008.
REFERENCES
- 1.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 983086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.al-Kobaisi, M. F., F. J. Rixon, I. McDougall, and V. G. Preston. 1991. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology 180380-388. [DOI] [PubMed] [Google Scholar]
- 3.Black, L. 1988. DNA packaging in dsDNA bacteriophages, p. 321-373. In R. Calendar (ed.), The bacteriophages, vol. 2. Plenum Press, New York, NY. [Google Scholar]
- 4.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 641007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowman, B. R., R. L. Welschhans, H. Jayaram, N. D. Stow, V. G. Preston, and F. A. Quiocho. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 802309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, J., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids, p. 111-155. In A. H. E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Acacdemic/Plenum Publishers, New York, NY.
- 7.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coller, K. E., J. I.-H. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 8111790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dasgupta, A., and D. W. Wilson. 1999. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J. Virol. 732006-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshmane, S. L., B. Raengsakulrach, J. F. Berson, and N. W. Fraser. 1995. The replicating intermediates of herpes simplex virus type 1 DNA are relatively short. J. Neurovirol. 1165-176. [DOI] [PubMed] [Google Scholar]
- 11.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2537-545. [DOI] [PubMed] [Google Scholar]
- 12.Fuller, D. N., D. M. Raymer, J. P. Rickgauer, R. M. Robertson, C. E. Catalano, D. L. Anderson, S. Grimes, and D. E. Smith. 2007. Measurements of single DNA molecule packaging dynamics in bacteriophage lambda reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 3731113-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gierasch, W. W., D. L. Zimmerman, S. L. Ward, T. K. Vanheyningen, J. D. Romine, and D. A. Leib. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135197-206. [DOI] [PubMed] [Google Scholar]
- 14.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7107-122. [DOI] [PubMed] [Google Scholar]
- 15.Homa, F. L., T. M. Otal, J. C. Glorioso, and M. Levine. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (γ2) gene lie within bases −34 to +124 relative to the 5′ terminus of the mRNA. Mol. Cell. Biol. 63652-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imber, R., A. Tsugita, M. Wurtz, and T. Hohn. 1980. Outer surface protein of bacteriophage lambda. J. Mol. Biol. 139277-295. [DOI] [PubMed] [Google Scholar]
- 17.Ishii, T., Y. Yamaguchi, and M. Yanagida. 1978. Binding of the structural protein soc to the head shell of bacteriophage T4. J. Mol. Biol. 120533-544. [DOI] [PubMed] [Google Scholar]
- 18.Jacobson, J. G., K. Yang, J. D. Baines, and F. L. Homa. 2006. Linker insertion mutations in the herpes simplex virus type 1 UL28 gene: effects on UL28 interaction with UL15 and UL33 and identification of a second-site mutation in the UL15 gene that suppresses a lethal UL28 mutation. J. Virol. 8012312-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, J. E., and W. Chiu. 2007. DNA packaging and delivery machines in tailed bacteriophages. Curr. Opin. Struct. Biol. 17237-243. [DOI] [PubMed] [Google Scholar]
- 20.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W.-H. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J. Virol. 724721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 722463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226403-407. [DOI] [PubMed] [Google Scholar]
- 23.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 721060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 806286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nixon, D. E., and M. A. McVoy. 2004. Dramatic effects of 2-bromo-5,6-dichloro-1-β-d-ribofuranosyl benzimidazole riboside on the genome structure, packaging, and egress of guinea pig cytomegalovirus. J. Virol. 781623-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 751427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel, A. H., and J. B. MacLean. 1995. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology 206465-478. [DOI] [PubMed] [Google Scholar]
- 28.Preston, V. G., J. Murray, C. M. Preston, I. M. McDougall, and N. D. Stow. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J. Virol. 826654-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 171030-1032. [DOI] [PubMed] [Google Scholar]
- 30.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 723779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Severini, A., A. R. Morgan, D. R. Tovell, and D. L. Tyrrell. 1994. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology 200428-435. [DOI] [PubMed] [Google Scholar]
- 32.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 7510755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tengelsen, L. A., N. E. Pederson, P. R. Shaver, M. W. Wathen, and F. L. Homa. 1993. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J. Virol. 673470-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 802118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 37.Trus, B. L., F. P. Booy, W. W. Newcomb, J. C. Brown, F. L. Homa, D. R. Thomsen, and A. C. Steven. 1996. The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol. 263447-462. [DOI] [PubMed] [Google Scholar]
- 38.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, K., and J. D. Baines. 2006. The putative terminase subunit of herpes simplex virus 1 encoded by UL28 is necessary and sufficient to mediate interaction between pUL15 and pUL33. J. Virol. 805733-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, K., F. Homa, and J. D. Baines. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 816419-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, Q., N. K. Maluf, and C. E. Catalano. 2008. Packaging of a unit-length viral genome: the role of nucleotides and the gpD decoration protein in stable nucleocapsid assembly in bacteriophage lambda. J. Mol. Biol. 381037-1048. [DOI] [PubMed] [Google Scholar]