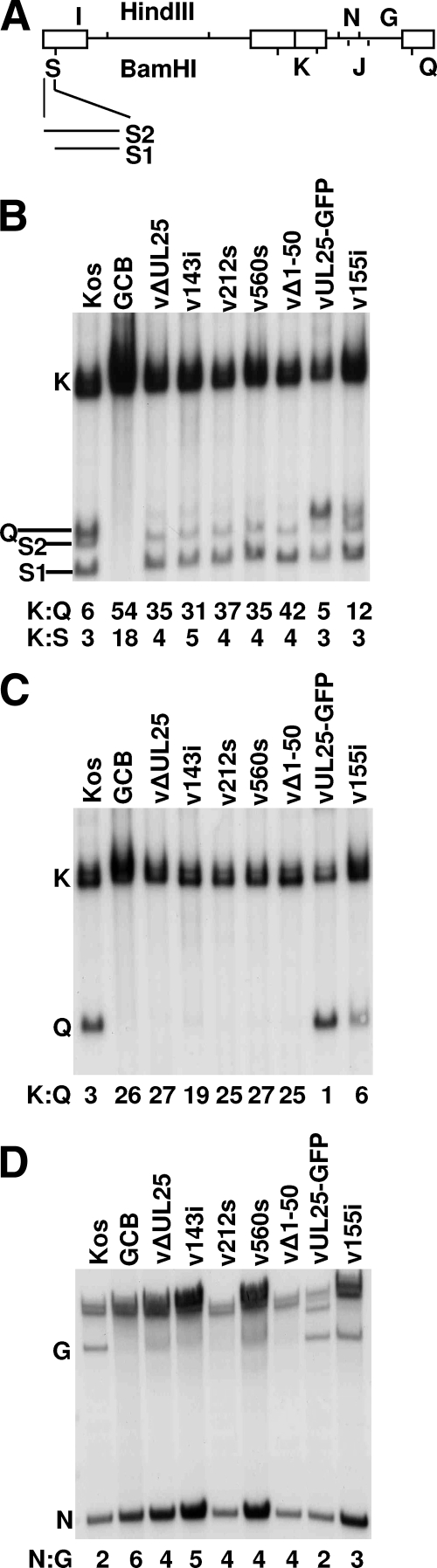
FIG. 7.
Processing of virus DNA. (A) Schematic diagram of the HSV genome showing the locations of the HindIII G, I, and N fragments and the BamHI J, K, Q, and S fragments. The different sizes of the BamHI S fragment are due to the presence of one to multiple copies of the a sequence at the UL terminus. (B to D) Vero cells were infected with the indicated virus at an MOI of 5 PFU per cell. At 18 h postinfection, total infected cell DNA was isolated, digested with BamHI (B and C) or HindIII (D), and subjected to Southern blot analysis. The blots were probed with 32P-labeled BamHI K (B), BamHI Q (C), or BamHI J (D) fragments of the HSV-1 genome. The ratio of the BamHI joint K fragment to US-end Q or UL-end S fragments and the ratio of the internal HindIII N fragment to the US-end G fragment were determined by quantification of the hybridizing bands with phosphorimaging software.