FIG. 1.
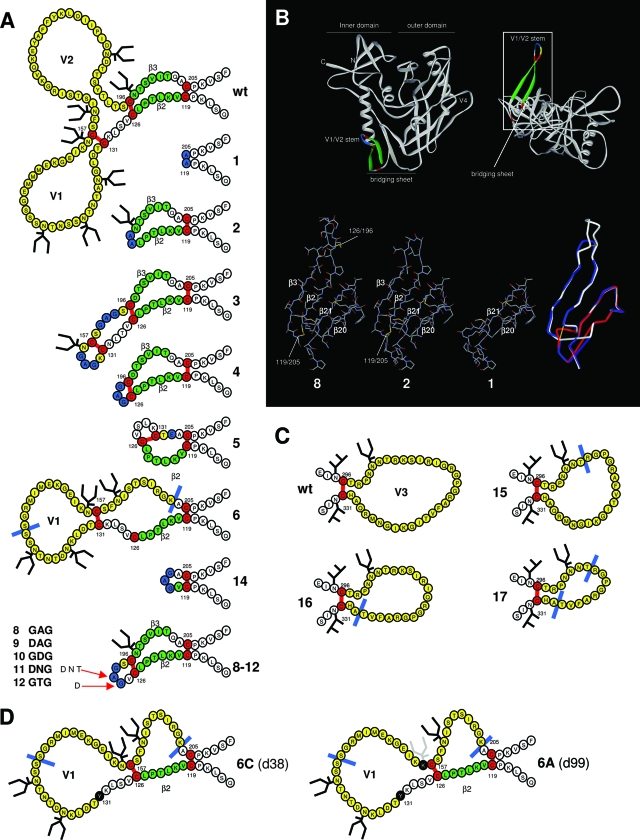
Design of the loop deletion variants. (A) Schematic representation of the V1/V2 deletion variants used in this study. The variable loops are indicated in yellow. The deletions are indicated by either a blue line or by blue-colored residues, which replace the deleted sequences. β-strands 2 and 3, components of the conserved bridging sheet, are indicated in green. Cysteines and disulfide bonds are colored in red. Note that the designation of the disulfide bonds is based on studies with the wt protein. We do not know whether the designated disulfide bonds do in fact form in these variants. This is particularly questionable in mutants 5 and 6 where one or two wt cysteine pairs cannot be formed. In variant 5, an alternative and hypothetical disulfide bond between 126 and 131 is drawn. In variant 6, the native C131-C157 bond is drawn and C126 is left unpaired. (B) Assumed 3-D models of selected ΔV1/V2 variants. The top panel provides perspectives on gp120 as seen from CD4 (left) and the coreceptor (right; rotated over the y axis by 90°). The rectangle in the top right panel encloses the V1/V2 stem and the bridging sheet. Colors are the same as described for panel A. The bottom panels represent details of this area for variants 1, 2, and 8 and an overlay of these variants (right bottom panel, variant 1 in red, variant 2 in blue, variant 8 in white, and disulfide bonds in yellow). The four β-strands that compose the bridging sheet and the local disulfide bonds are indicated. The LAI gp120 core and variant cores were modeled by SWISS-MODEL (http://swissmodel.expasy.org//SWISS-MODEL.html) (26) using the HXB2 core (pdb accession code 1G9M [37]) and drawn using Viewerlite (Accelrys, Inc.). The overlay in the bottom right panel was prepared with Deepview/SWISS pdb viewer (http://www.expasy.org/spdbv/) (26) and rendered in Viewerlite. (C) Schematic representation of the V3 deletion variants. Colors are as described for panel A. (D) Rearrangement of the V1/V2 stem in variant 6. The starting situation is as described for panel A. Note that the drawn disulfide bond between residues C131 and C157 is purely speculative. However, in the wt protein, these cysteines do form a disulfide bond. Left panel, a hypothetical situation after the first substitution (C131Y) with a new nonnative disulfide bond between C126 and C157, resulting in the restoration of the V1 to its full length and the formation of a pseudo-V2. Right panel, removal of N156 after prolonged culturing. Note that we observe the removal of the glycosylation site at N156 in two independent cultures in two different substitutions, N156K (as indicated in the figure) in culture 6A and S158F in culture 6B. The sequences were derived from sequencing clones at day 38 (6C) and day 99 (6A).