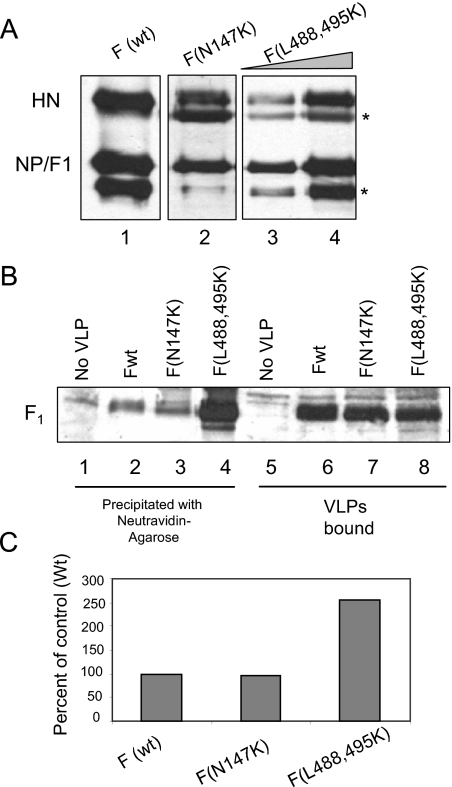
FIG. 5.
MPB labeling of F mutants in VLPs. (A) VLPs were generated using Fwt (lane 1) or HR1 mutant, F (N147K) (lane 2) or HR2 mutant, F (L488,495K) (lanes 3 and 4). The amounts of VLPs generated in each case were estimated by detecting particle associated proteins by a Western blot using a mix of anti-NDV antibody, anti-F antibody, and anti-HN (anti-AS) antibody. The protein band below the NP protein (*) is a degradation product of NP variably seen in different preparations of virus and VLPs (unpublished observation). The protein band below the HN protein (*) is a nonspecific protein band variably detected by anti-AS antibody, an anti-HN antibody. (B) Equivalent amounts of VLPs (normalized to NP protein shown in panel A) with Fwt (lanes 2 and 6) or HR1 mutant F (N147K) (lanes 3 and 7) or HR2 mutant F (L488,495K) (lanes 4 and 8) were incubated with RBCs and MPB at 4°C for 1 h, followed by 22°C for 15 min. RBCs incubated with MPB in the absence of VLPs (lanes 1 and 5) were used as a control. Unbound VLPs and MPB were removed, and F protein in bound VLPs was analyzed for MPB labeling by precipitation with neutravidin-agarose (lanes 1 to 4), followed by Western analysis with anti-HR2 antibody. Extracts from RBCs (lanes 5 to 8) were resolved as a control for the amount of VLPs bound to RBCs. (C) Quantification of MPB labeling in panel B. The results are representative of two similar experiments. Quantification was accomplished using blots exposed in a linear range of film. As shown in Fig. 1 to 4 and in other experiments, there was no nonspecific precipitation of wild-type or mutant F protein with neutravidin-agarose in the absence of MPB binding.