Abstract
Nudaurelia capensis ω virus-like particles have been characterized as a 480-Å procapsid and a 410-Å capsid, both with T=4 quasisymmetry. Procapsids transition to capsids when pH is lowered from 7.6 to 5.0. Capsids undergo autoproteolysis at residue 570, generating the 74-residue C-terminal polypeptide that remains with the particle. Here we show that the particle size becomes smaller under conditions between pH 6.8 and 6.0 without activating cleavage and that the particle remains at an intermediate size when the pH is carefully maintained. At pH 5.8, cleavage is very slow, becoming detectable only after 9 h. The optimum pH for cleavage is 5.0 (half-life, ∼30 min), with a significant reduction in the cleavage rate at pH values below 5. We also show that lowering the pH is required only to make the virus particles compact and to presumably form the active site for autoproteolysis but not for the chemistry of cleavage. The cleavage reaction proceeds at pH 7.0 after ∼10% of the subunits cleave at pH 5.0. Employing the virion crystal structure for reference, we investigated the role of electrostatic repulsion of acidic residues in the pH-dependent large conformational changes. Three mutations of Glu to Gln that formed procapsids showed three different phenotypes on maturation. One, close to the threefold and quasithreefold symmetry axes and far from the cleavage site, did not mature at pH 5, and electron cryomicroscopy reconstruction showed that it was intermediate in size between those of the procapsid and capsid; one near the cleavage site exhibited a wild-type phenotype; and a third, far from the cleavage site, resulted in cleavage of 50% of the subunits after 4 h, suggesting quasiequivalent specificity of the mutation.
Virus maturation (i.e., the transition from noninfectious particle to infectious virion) is common in complex virus formation. In general, initial assembly occurs with tenuous contacts between subunits so that the process can be self-correcting in a time course that places all the subunits in the proper location. The resulting particle, often called a procapsid, is often fragile and not infectious. Maturation to a robust infectious virion often involves large conformational changes (LCCs) and autocatalytic chemistry occurring within the particle. Only a limited number of model systems, mostly in the bacteriophage family, are sufficiently accessible for biophysical and biochemical analysis (24). Here we investigate the LCC and autoproteolytic chemistry associated with maturation of the nonenveloped tetraviruses.
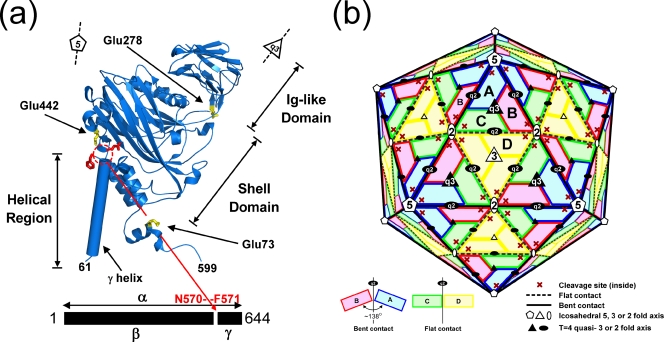
Tetraviruses are T=4, nonenveloped, icosahedral viruses that possess single-stranded, positive-sense RNA genomes (15, 23). The Tetraviridae family is subdivided into the omegatetraviridae genus and the betatetraviridae genus. Nudaurelia capensis omega virus (NωV) infects insects and can be grown only in whole Lepidoptera larvae. NωV copackages a bipartite genome with a ∼5.3-kb RNA1 that codes for the RNA-dependent RNA polymerase and a ∼2.5-kb RNA2 that encodes the 644-residue coat protein (2, 14). NωV has a maximum diameter of 430 Å, with an internal sphere of diameter 235 Å occupied by the RNA (VIPERdb; http://viperdb.scripps.edu/) (22). Investigation of the crystal structure of NωV (16, 20) revealed that the four subunits in the icosahedral asymmetric unit (i.e., 240 subunits in the particle) had very similar tertiary structures formed by an internal helical domain, a shell domain with the classic virus jelly-roll fold, and an external domain with an immunoglobulin (Ig) fold. The latter is inserted between two strands of the jelly roll, and the helical domain has contributions from both the N- and C-terminal regions of the subunit. The crystal structure revealed that the capsid protein in all four subunits had undergone a proteolytic cleavage between residues 570 and 571, with the 74-residue polypeptide forming part of the inner helical domain but with configurations dependent on the subunit location in the quaternary structure. The sequence of the capsid protein gene indicated that the initial gene product would be the full 644 residues (called alpha); following cleavage, the products were termed beta (residues 1 to 570) and gamma (residues 571 to 644) (Fig. 1a). The subunits were arranged as predicted by Caspar and Klug (9), with A subunits forming pentamers and pairs of B, C, and D subunits forming quasihexamers at the icosahedral twofold axes. A, B, and C subunits form quasithreefold interactions, while D subunits are related by icosahedral threefold rotations. There are two types of quasitwofold contacts, those connecting triangles containing the A, B, and C subunits and those connecting a triangle with A, B, and C subunits to a triangle containing D subunits (Fig. 1b). While threefold and quasithreefold interactions are closely similar, the two types of quasitwofold contacts are dramatically different. Quasitwofold interactions between ABC triangles form a dihedral angle (the angle between the planes defined by the two triangles) of ∼138°, while the angle between the ABC and DDD triangles is ∼180° (Fig. 1b). The difference is remarkable, because the interfaces contain the same subunit surfaces; however, in the former case (bent contact), the surfaces directly interact with each other, while in the latter case (flat contact), the interaction is mediated by a helical polypeptide (a portion of gamma from subunits C and D) that prevents bending at lines of interaction (the hinges) that are equivalent at both types of quasitwofold. The subunits rotate about the hinge essentially as a rigid body, generating close contacts between the helical domain and a wide separation between Ig-like domains at the bent contact, while the helical domains are separated and the Ig-like domains are closely associated at the flat contact.
FIG. 1.
(a) The structure of the A subunit determined by the crystallographic analysis and refinement of NωV. The autoproteolytic cleavage occurs between Asn570 and Phe571 (shown as red stick side chains). The locations of three mutations discussed in the manuscript (Glu73, Glu278, and Glu442) are shown as yellow stick side chains. The structures of the B, C, and D subunits are closely similar to that of the A subunit, with some variations in the helical domain. (b) A schematic drawing of the T=4 subunit arrangement in the NωV particle. The quasiequivalent A, B, C, and D subunits are shown in blue, pink, green, and yellow, respectively. This color coding is maintained in all the figures that follow. Icosahedral and quasirotational axes are shown with white and black symbols, respectively. Note that the icosahedral twofold axes are quasisixfold axes in this surface lattice. Subunit contacts at the A/B interface (bent) have a dihedral angle of ∼138° (solid black line in the particle) and at the C/D interface (flat) have a dihedral angle of 180° (broken line).
The lack of a cell line to propagate NωV made it impossible to study particle assembly and maturation. Expression of the NωV capsid protein gene in the baculovirus system, however, produced virus-like-particles that had the same dimension in negative-stained electron micrographs as the authentic virus and had undergone proteolysis to generate the gamma peptide (1). These particles contained predominantly cellular RNA that was apparently nonspecifically packaged during assembly. The A260/280 ratio for these particles was 1.2, comparable to that of virions. Subsequently, the purification of virus-like particles (VLPs) was modified, employing a buffer at pH 7.6 instead of the pH 5.0 buffer originally used. These particles had not undergone proteolysis and had a larger dimension (diameter, 480 Å) than authentic NωV (6). The electron cryomicroscopy (cryoEM) analysis of these VLPs showed that they were T=4 quasisymmetric, with large pores at fivefold and twofold (quasisixfold) axes and a morphology entirely different from that of authentic virions, dominated by dimer contacts on the outer surface and trimer contacts on the inner surface. The observation that the two classes of dimer contact were similar led to the suggestion that the dimer is preformed in solution and is the assembling unit during particle formation. Lowering the pH from 7.6 to 5, in vitro, reduced the particle size, changed the surface morphology, and induced autoproteolytic cleavage, all of which resulted in the formation of particles that were indistinguishable from authentic virions or from the VLPs purified at pH 5.0. The A260/280 ratio was 1.2 before and after the LCC, indicating no loss of nucleic acid during the transition. Small-angle-scattering studies combined with stop-flow kinetic analysis revealed that the LCC occurred in less than 100 ms at pH 5.0 (7). However, the cleavage occurred at a slow rate, requiring hours for completion. In the same investigation it was shown that the LCC was reversible when the pH was returned to 7.0 before ∼15% of the subunits cleaved. Taylor et al. showed that a particle with a noncleaving mutation (Asn570Thr) retained reversibility indefinitely (26), indicating that the cleavage allowed interactions that stabilized the compact form even at pH 7.0 or above.
Here we demonstrate that through titration, the particle dimension can be changed from that of a procapsid to that of a capsid in a continuous manner, with intermediate sizes maintained indefinitely when the pH is carefully controlled. At pH 5.8, the particle has the dimension of the capsid, but cleavage is detected only after ∼24 h. At pH 5.0, cleavage is optimal, with a half-life of 30 min. Particles acquiring ∼10% of the subunits cleaved at pH 5 can be shifted to pH 7, under which conditions the cleavage proceeds with kinetics comparable to that seen at pH 5, showing that low pH is required for the conformational change and, presumably, for formation of active sites but not for the chemistry of cleavage. The titration behavior and calculation of pKa values of acidic residues based on the crystal structure suggest that maturation is controlled by the protonation of Asp and Glu residues that dominate subunit interfaces. The details of the contributions of residues deemed significant on the basis of analysis of the structure were explored with Glu-to-Gln mutations. Three of these assembled properly, and their biophysical and biochemical properties were characterized. Three different phenotypes were found for these particles and were interpreted in terms of the structure.
MATERIALS AND METHODS
Construction, expression, and purification of NωV wild-type and mutant VLPs.
The coding region of the NωV coat protein in pBackPAC-9 baculovirus transfer vector (25) was inserted into ProGreen vector (AB Vector) between 5′-BamHI and 3′-XbaI restriction sites. The ProGreen vector is identical to the parental BacPAK-9 vector except that it provides for the expression of the Aequorea victoria green fluorescent protein. The mutated gene was generated from the transfer vector by the use of the quick-change mutagenesis method. The procedure from plaque purification to infection was followed using the protocol of the manufacturer (GibcoBRL). Expression of coat proteins was confirmed by Western blotting using the polyclonal anti-NωV coat protein antibody. Purification of VLPs was performed as previously described (25). All VLPs were purified at pH 7.6 as the procapsid form and then concentrated to 1.1 mg/ml in 10 mM Tris-HCl (pH 7.6)-250 mM NaCl-5 mM EDTA.
Velocity sucrose gradient analysis.
The Asn570Thr mutant, which is cleavage defective, was expressed and purified as described previously (26). Asn570Thr mutants were sedimented on a 10-to-30% sucrose gradient prepared at different pH values. Each gradient contained the same ingredients except for the buffer, which was chosen for maximum stabilization of the pH and consisted of the following ingredients: 250 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl (for pH 7.6), and 50 mM Mes/NaOH (for pH 6.0 or 6.8) or 50 mM sodium acetate (for pH 5.0). Biocomp Gradient Master was employed for preparing the 10-to-30% sucrose gradients at the indicated pH values. The sample was centrifuged using a Beckman SW41 rotor at 40,000 rpm for 1.25 h at 11°C and then fractionated on an ISCO fractionator equipped with an absorbance detector (UA-6). All data were normalized by elution volume and time. Note that particles were maintained at pH 7.6 and then placed on the gradient at the pH indicated and centrifuged. Thus, in each case, the particles were at the lower pH for 1.25 h.
Autocatalytic cleavage assays.
The reactions were performed by mixing 1 volume of concentrated NωV procapsids in 10 mM Tris-HCl buffer and 9 times the volume of the designated lower-pH buffer (100 mM sodium acetate, 250 mM NaCl, and 5 mM EDTA). All reactions were incubated at 20 to 21°C and then stopped by adding the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and by immediately freezing with liquid nitrogen. The SDS-PAGE analysis (NuPAGE 4-to-12% Bis-Tris gel; Invitrogen) was employed to estimate both alpha and beta peptide quantities after cleavage. All frozen samples were quickly heated in boiling water for 5 min before loading. The gels were stained, digitized, and analyzed with the program ImageJ (http://rsb.info.nih.gov/ij/). The data represented the means (± standard deviations) of the results of three independent measurements.
Autocatalytic cleavage assays at neutral pH.
The reactions were performed by mixing 1 volume of concentrated procapsid in 10 mM Tris-HCl buffer and 1.5 times the volume of lower-pH buffer (50 mM sodium acetate, 250 mM NaCl, 5 mM EDTA). At designated times (at 1 min or during the time course), the 6× volume of pH 7.6 buffer (100 mM Tris-HCl, 250 mM NaCl, 5 mM EDTA) was added into 1 volume of the reaction mixture and then the combined volumes were incubated for 9 h. The final pH value was confirmed to be 7.0 by performing the dilutions without VLPs in a volume sufficient to directly measure the pH. The reactions were also stopped, applied to the gel that was digitized, and analyzed as described above. The results represented the means (± standard deviations) of the results of three independent measurements.
Calculation of the electrostatic surface and protein pKa values.
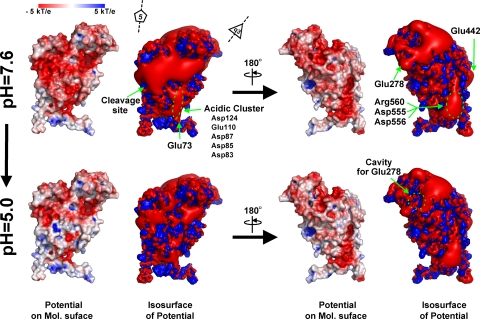
All the following calculations of the electrostatic surface and protein pKa values were performed with the three-dimensional coordinates using PDB2PQR/ProPKA software packages and the Adaptive Poisson-Boltzmann Solver software package (3, 11, 12, 17). The Protein Data Bank identification of 1OHF was employed for the three-dimensional coordinates of the NωV capsid form, and the previous pseudoatomic model of the procapsid form was also employed for the calculations (6, 16). First, an isolated A subunit was used to assign protein titration states and to calculate the electrostatic potential as a monomeric soluble protein, and then the resulting potential was illustrated using Pymol software (http://www.pymol.org) (see Fig. 5). Note that the results at pH 7.6 and 5.0 were calculated without the environment due to neighboring subunits in order to illustrate the electrostatic potentials; the low-level dielectric environment due to the presence of neighboring proteins substantially perturbs the local electrostatic potential by accentuating regions of strong positive and negative potential. Second, the protein pKa values of each of the four quasiequivalent subunits were calculated with those of the surrounding subunits of above procapsid or capsid three-dimensional coordinate (Table 1). A complete list of the pKa values for all of the ionizable residues is available from the authors. All graphical representations in the present paper were produced using Pymol and UCSF Chimera software (http://www.cgl.ucsf.edu/chimera/) (21).
FIG. 5.
Electrostatic potential of an isolated A subunit at pH 7.6 and 5.0; the molecular surface is colored by ±5-kT/e potentials calculated at the solvent-accessible surface and shown as blue and red gradations, respectively. Also, the ±3-kT/e potential isocontours are shown as blue and red surfaces, respectively. The view of both representations (left four panels) corresponds to Fig. 1a. The electrostatic potential was calculated for the monomeric protein, meaning that these potentials are not be influenced by the low-level dielectric environment or the charge distribution of neighboring subunits.
TABLE 1.
Results of mutations in the NwV coat protein
| Mutation(s) | Assembly | LCC | Proteolytic kineticsa | Residual pKab
|
|
|---|---|---|---|---|---|
| Procapsid | Capsid | ||||
| Glu73Gln | Yes | Native-like | Aberrant | (Glu73) 2.44-5.24 | (Glu73) 1.74-3.92 |
| Asp124Asn | No | (Asp124) 2.4-4.44 | (Asp124) 1.98-8.10 | ||
| Glu278Gln | Yes | Native-like | Aberrant | (Glu278) 4.5 | (Glu278) 6.27-6.73 |
| Glu442Gln | Yes | Native-like | Native-like | (Glu442) 5.89-6.40 | (Glu442) 8.03-9.58 |
| Asp87Asn/Asp124Asn | No | (Asp87) 2.22-9.31 | (Asp87) 6.49-12.84 | ||
See Fig. 7.
A complete list of the pKa values for all ionizable residues is available from the authors.
CryoEM analysis and model building of the Glu278Gln mutant.
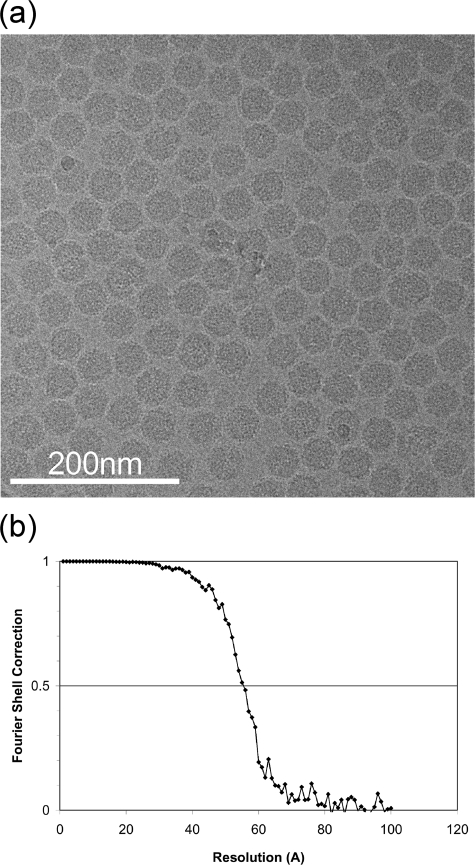
Purified Glu278Gln VLPs were concentrated to ∼40 mg/ml in 50 mM Tris-HCl (pH 7.6)-250 mM NaCl. One volume of the concentrated sample was then mixed with 1 volume of pH 4.7 buffer (100 mM sodium acetate, 250 mM NaCl). The final pH of the reaction mixture was independently confirmed to be 5.0. Three microliters of the reaction solution was placed on the grid and flash frozen using a Vitrobot specimen preparation unit (FEI). The data set was collected using an FEI Tecnai F20 electron microscope operated at 120 keV at liquid-nitrogen temperature. Data were collected at 1.0-to-3.0 microns under focus, and the instrument was controlled by use of LEGINON software (8). The contrast transfer function of each micrograph was estimated and phase-corrected using the program ACE (19). A total of 27,386 particles were collected by use of the program SELEXO from 462 images that were manually selected (27). Those particles were centered in 200-by-200-pixel boxes, corresponding to 2.768 Å/pixel. Single-particle three-dimensional reconstruction refinement was carried out with the EMAN software suite (18), using the capsid crystal structure as the initial model. Icosahedral symmetry was imposed during the reconstruction refinement, resulting in the final 9.8-Å resolution structure as assessed by the 0.5 Fourier shell correlation criteria. The program SFALL in the CCP4 suite (10) was used to generate a list of structure factors from the resulting density reconstruction. Coordinates of the N- and C-terminal regions were truncated from the X-ray model, with the remaining residues refined as rigid bodies in the four quasiequivalent subunits. The residues used for the A, B, C, and D subunit refinement procedures were residues 79 to 589, 79 to 589, 78 to 589, and 78 to 589, respectively. The rigid-body refinement of the model into reconstructed density values was performed with UCSF Chimera (21) and CNS (5) software. See Fig. 9 for a typical micrograph and the Fourier shell correlation curve.
FIG. 9.
(a) A typical micrograph of the Glu278Gln mutant particles flash frozen and imaged at ×80,000 magnification. (b) Resolution determination of the reconstruction based on the Fourier shell correlation method. The particle images used for the reconstruction refinement were divided into equally sized data sets, and two new reconstructions were calculated. The correlation coefficients of the two resulting Fourier shell reconstructions were then computed to estimate the resolution. The 0.5 Fourier shell correlation criterion was 9.8 Å.
RESULTS
Particles decrease in size (LCC occurs) between pH 7.6 and 6.0 in a continuous manner.
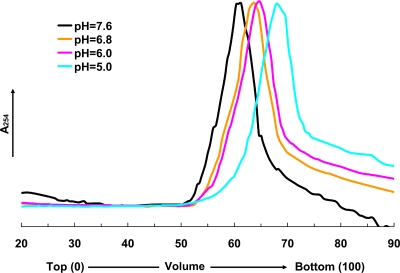
The pH dependence of the LCC change was examined using the Asn570Thr (noncleaving) mutant with velocity sucrose gradient (10 to 30%) analysis under conditions between pH 7.6 and 5.0 (Fig. 2) to compare with similar studies performed with the wild-type particle by use of small-angle X-ray scattering (7). The noncleaving mutant was used to eliminate any effect of cleavage on the LCC change. At pH values of 6.8 and 6.0, based on sedimentation velocity, particles of intermediate sizes between those of the procapsid (pH 7.6) and capsid (pH 5.0) were observed. They maintained the same dimension for ∼3 h, indicating that the LCC probably occurs in a continuous manner in this pH range and that the size may be proportional to the degree of protonation of the acidic residues. Under conditions below pH 6.0, it was not possible to observe additional intermediate particle sizes, suggesting that the latter stages of the transition cannot be controlled for the 2-to-3-h time period required for measurement of the sedimentation rate of the particles. In addition, the change in dimension is smaller in this range and the gradient may not have been capable of resolving such small changes. From this analysis, we were able to distinguish only two particles of intermediate size with confidence; however, it is possible that the size transition occurs discontinuously at lower pH.
FIG. 2.
The velocity sucrose gradient (10 to 30%) analysis of the cleavage-defective Asn570Thr mutant. The analysis was performed by placing particles initially at pH 7.6 onto gradients between pH 7.6 and 5.0. Centrifugation at 40,000 rpm was performed immediately and maintained for 1.25 h (corresponding to the incubation time at the designated pH). Peaks in the gradients at pH 7.6 and 5.0 correspond to the procapsid and capsid velocities, respectively. The sedimentation velocities at pH 6.8 and 6.0 indicate intermediate particle sizes. Between pH 6.0 and 5.0, it was impossible to detect differences in the sedimentation velocity of the particles even though, in the wild type, the proteolytic cleavage was not detected at pH 6.0 and proceeded relatively rapidly (see below) at pH 5. The latter observation indicates that there is a difference in the extent of the LCC at these two pH values; however, the experiment had insufficient resolution to detect this.
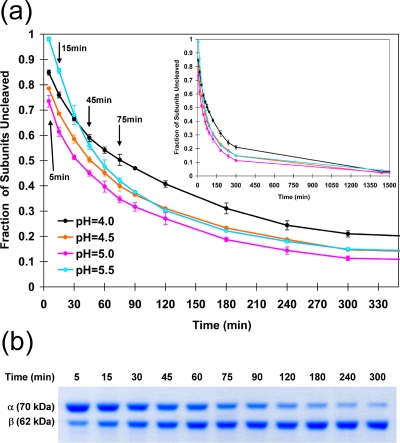
Maturation cleavage occurs at a maximal initial rate at pH 5.0 (half-life, ∼30 min).
The kinetics of cleavage was examined under conditions between pH 4.0 and 5.5 (Fig. 3). The initial rate of cleavage was maximal at pH 5.0, while the latter stages of cleavage (i.e., after ∼50% of the subunits had cleaved) were less sensitive to pH. At a pH value of 6.0, cleavage was not detectable. At pH 5.8, there was no initial cleavage; however, after 24 h, some cleavage could be detected (data not shown). Indeed, the cleavage rate even at pH 5.5 was initially extremely slow. At optimum pH values, cleavage occurred rapidly; however, after 24 h there was always a residual of uncleaved subunits, amounting to approximately 5% (Fig. 3).
FIG. 3.
(a) Kinetics of cleavage measured for wild-type VLPs at different pH values. VLP procapsids were exposed to the designated pH, and aliquots were frozen in liquid nitrogen at the time points indicated. SDS gel analysis was then performed, and the fraction of subunits cleaved was determined by optical scanning of the gels to determine the amount of alpha and beta at the time point. The maximum initial rate was measured for particles incubated at pH 5. (b) A typical SDS-PAGE gel (wild-type NωV) under conditions of a time course incubation at pH 5.0.
Autoproteolysis can proceed at neutral pH after LCC at pH 5.0.
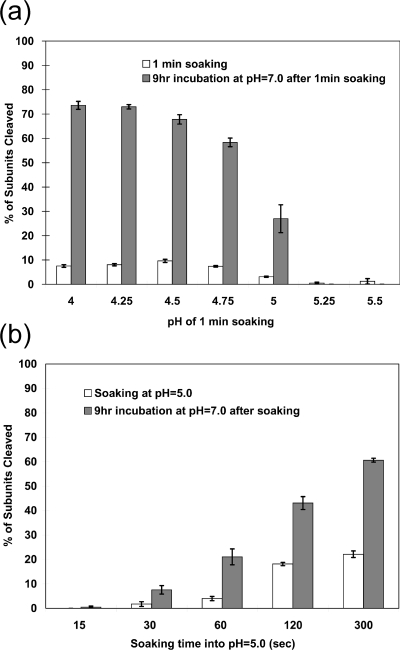
Experiments were performed to examine the effect of pH on the chemistry of the autocatalytic cleavage. Samples were exposed to pH values between 5.5 and 4.0 for 1 min and then raised to pH 7.0. The extent of cleavage was measured after 1 min and after 9 h (Fig. 4a). Between pH 4.0 and 4.75 all the samples showed approximately 10% cleavage at 1 min. All of these samples were ∼60% cleaved after 9 h at pH 7.0, indicating that the cleavage occurs efficiently at neutral pH. In contrast, samples incubated at pH values above 5.0 showed little initial cleavage and no additional cleavage at pH 7.0. A time course experiment was then performed at pH 5.0 in which samples were incubated for between 15 and 300 s and then shifted to pH 7.0 for 9 h (Fig. 4b). There was a clear correlation between the soaking time at pH 5.0 and the total amount of cleavage that occurred after 9 h. As elaborated in the Discussion, our results show that conditions of low pH are required for the LCC necessary to form the active site for cleavage but that after ∼10% of the subunits cleave, the reaction is independent of pH level.
FIG. 4.
Autoproteolysis occurs at neutral pH after activation at low pH. (a) The progress of the autoproteolytic cleavage at neutral pH values following activation at lower pH. The wild-type VLP procapsids were soaked in buffers between pH 4.00 and 5.50 for 1.0 min. The pH was then immediately shifted to pH 7.0, and the particles were incubated for 9.0 h. The fractions of subunits cleaved at 1 min and 9 h were determined as described for Fig. 3. (b) The progress of the autoproteolytic cleavage at neutral pH values following incubation at pH 5.0 for between 15 and 300 s. After being soaked at the indicated time points, the pH values were immediately changed to pH 7.0 and the mixture was incubated for 9 h. The chemistry of the reaction does not require low pH. The data indicate that ∼10% of the subunits must initially cleave at low pH for the reaction to proceed at pH 7.0, a result consistent with the observations of Canady et al. (7). We conclude that when fewer subunits cleave, the LCC is reversible. Panel b shows a relationship between the amount of initial cleavage and that seen after the reaction proceeds, suggesting that acidic residues with pKa values above 5.0 would play a key role in the LCC at low pH values.
Acidic residues at subunit interfaces create electrostatic repulsion at neutral pH.
Figure 5 shows the computed electrostatic surface potential, based on the crystallographic coordinates, of an isolated A subunit (i.e., the environment due to neighboring subunits is not included in the pKa calculations). In addition to an overall negative charge at pH 7.0, there are clear regions of concentrated negative charge. When the same calculation is performed for the capsid proteins, including their neighbor environment in the calculations, the pKa values of many of the acidic residues rise by 2 to 3 pH units, indicating that protonation would occur at a pH that is higher than expected. Employing the pseudoatomic model of the procapsid for the same calculation revealed that, even in this state, a number of acidic residues have elevated pKa values. We propose that protonation of these residues in the procapsid at pH 6.8 initiates the transition (Fig. 2) and that each stage of particle contraction progressively raises the pKa of some acidic residues, making the transition complete at pH 5.0. The progressive elevation of pKa values on contraction explains why the LCC occurs at pH 5.0 and not at the pKa of normal acidic residues (pH ∼ 4). The outer-surface morphology of the procapsid particle is dominated by dimer contacts (6), suggesting that the major repulsion occurs between trimer interfaces at pH 7.6. At pH 5.0, these have been protonated and the capsid surface displays predominantly trimer morphological units. The Ig-like domains of the subunit dominate the surface, but since the subunits move mostly as rigid bodies, they represent the overall change in subunit contacts during the transition. The role of acidic residues deemed significant in the transition, based on the structure and potential changes in pKa values, was examined with site-directed mutagenesis.
The mutations Asp124Asn and Asp87Asn/Asp124Asn are expressed, but expression does not result in the formation of particles.
These mutations (Table 1) are located at the two classes of quasitwofold axes (flat and bent in the mature capsid) and are associated with a large cluster of acidic residues on the lower portion of the shell domain (Fig. 5). These clusters are related by twofold symmetry at the hinge that pivots to allow formation of either the flat or bent contact. His541is located next to the cluster. His541 in the B subunit (pKa > 7 in the procapsid and capsid) forms salt bridges with the cluster of neighboring A subunit (bent contact) in the capsid. The phenotype of these mutations clearly indicates that this sensitive portion of the structure does not tolerate even subtle changes. Indeed, as noted in the introduction, the dimer is probably the assembling unit; when it cannot form, assembly is aborted.
The mutation Glu442Gln near the cleavage site has the wild-type phenotype for LCC and cleavage kinetics.
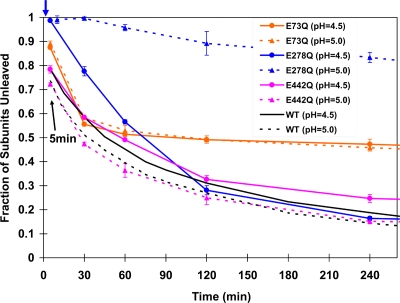
This residue was chosen for mutation because of its exceptionally high pKa in the capsid, its close proximity to the cleavage site (on a neighboring polypeptide chain), and its association with icosahedral fivefold and quasisixfold particle axes (Fig. 5). Removing the pH dependence of this residue was expected to affect both the LCC and the cleavage. In fact, there was no discernible change from wild type in the parameters measured (Fig. 6).
FIG. 6.
The cleavage assay of mutants at pH 5.0 and 4.5. The data for the wild type (Fig. 3a) are shown in black for comparison. An arrow at the top indicates the time point (2 min) at which Glu278Gln VLPs at pH 5.0 were frozen for cryoEM analysis.
The mutation Glu73Gln allows only 50% of the subunits to cleave in 4 h; wild-type particles are 85% cleaved in the same period.
Glu73 was chosen for mutation because it is located on the inner helical domain, forms a salt bridge with Arg560 (Fig. 7) that is surrounded by acidic residues on the same chain, and had a normal pKa value in the capsid (Fig. 5), suggesting that it was charged even after the LCC. We anticipated that interruption of the salt link would have a subtle effect on the subunit interface at this position, with a possible effect on the LCC. The phenotype was remarkably clear, with cleavage virtually stopped after 50% of the subunits were processed, although the initial cleavage rate was comparable to that of the wild type (Fig. 6). Clearly, the LCC was not affected by the mutation; however, subtle effects must have been induced at two of the four cleavage sites in the particle, inhibiting proteolysis in these subunits.
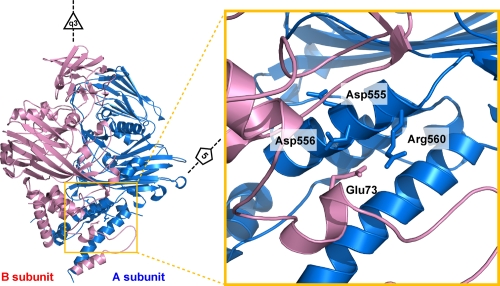
FIG. 7.
A closeup view in the region near Glu73 (B subunit; shown in pink). Glu73 is located at the interface of subunits related by icosahedral and quasithreefold symmetry and occupies virtually identical positions in all four subunits. It forms a salt bridge with the neighboring Arg560 (A subunit; shown in blue) that is surrounded by negative charges (Arg560 and Asp556), as shown in Fig. 5. The phenotype of this mutation was unexpected, because only half the subunits cleave at pH 5.0 and 4.5, indicating an effect of quasiequivalence, and yet the residue displays high-fidelity quasiequivalence.
The mutation Glu278Gln at the threefold axes and far from the cleavage site produces an unexpected phenotype: inhibition of the LCC at pH 5.0.
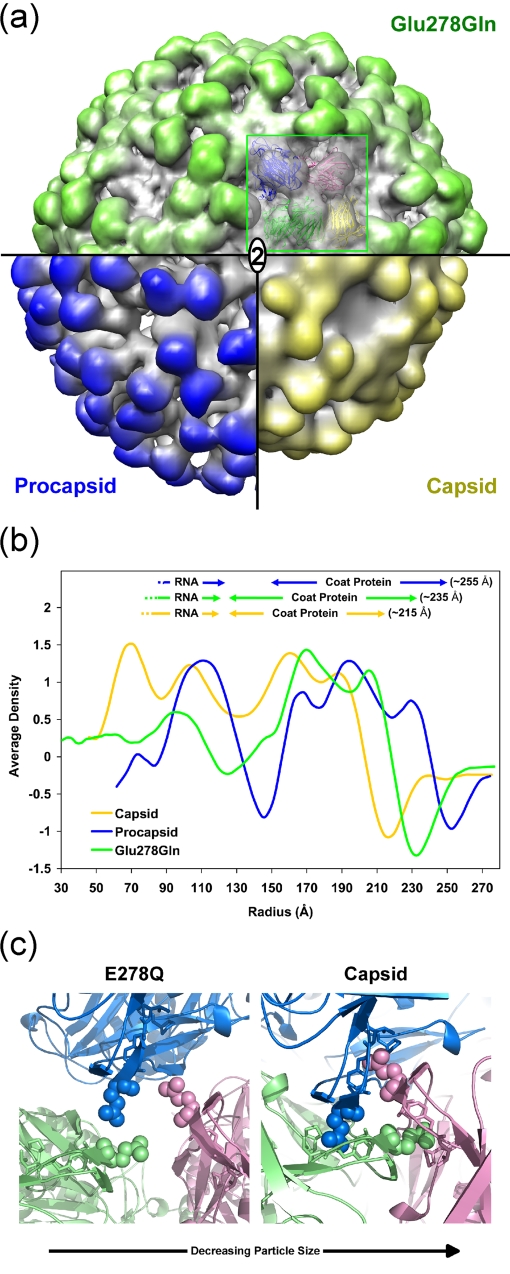
Glutamic acid 278 was selected for mutation because it changed from a normal pKa value (4.5) in the procapsid to 6.3 in the capsid and because it is on the polypeptide connecting the Ig-like domain to the shell domain far from the cleavage site. It is close to the icosahedral and quasithreefold axes, and in the capsid, symmetry-equivalent Glu278 residues are in close proximity. We anticipated that changing the Glu to Gln would reduce electrostatic repulsion and allow the LCC to occur at higher pH. In fact, at pH 5.0, there was no cleavage in the first 30 min, and pH values of at least 4.5 were required to initiate cleavage in the first 30 min (Fig. 7). This suggested that the mutation inhibited the LCC at pH 5.0; this was confirmed by the cryoEM reconstruction at 9.8-Å resolution (Fig. 8a). Figure 8b shows that this mutant is intermediate in size between the wild-type procapsid and capsid. The rationale for this behavior is discussed below.
FIG. 8.
(a) A surface volume representation of the Glu278Gln mutant at 9.8-Å resolution (green), wild-type capsid at 25 Å resolution (yellow), and wild-type procapsid at 28-Å resolution (blue) (6). A pseudoatomic model of the mutant particle was generated by fitting the atomic coordinates, as a rigid body, to the observed density values. 2, twofold axis. (b) Comparison of the Glu278Gln mutant capsid, wild-type capsid, and wild-type procapsid (6) radially averaged electron density distributions. The averaged outer edge of the mutant corresponded to an intermediate size (∼235 Å) compared to those of the procapsid (∼255 Å) and capsid (∼215 Å). (c) A view of the quasithreefold axis near Glu278. At the left is a closeup view of the Gln278 pseudoatomic model described for panel a at pH 5.0. Glu278 is shown in the crystal structure of the capsid at the right. In the capsid form, Glu278 is inserted into a cavity formed in the neighboring subunit. The cavity consists of residues Ala276, Phe282, Val283, and Pro489 (side chains are shown as stick models). These hydrophobic residues are surrounded by negative charges at pH 7.6; however, the charge is reduced at pH 5.0 (Fig. 5). The LCC in the Glu278Gln particle was inhibited by this substitution, indicating that the self-repulsion between symmetry-related Glu278 residues plays a crucial role in the cavity insertion.
DISCUSSION
Figure 4 shows that the chemistry of the cleavage reaction does not depend on pH values near 5.0. Particles were exposed to pH values below 5.5 for 1 min and then raised to pH 7.0. At pH values below 5.0 approximately 10% of the subunits underwent cleavage in the first minute; however, more than 60% of the subunits cleaved during a 9-h incubation at pH 7.0. The data are consistent with previous observations in which small-angle-scattering measurements showed that particles were locked in the capsid form after ∼15% of the subunits were cleaved (7). When the pH was raised to 7.0 before that point, particles expanded to an intermediate size between those of the capsid and procapsid. Data presented here show that the number of cleaved subunits required to retain the capsid form at pH 7.0 is approximately 5% and that capsids in this form are capable of autocatalytic proteolysis at neutral pH. Thus, pH values below 5.0 are required to induce and stabilize the LCC and presumably to form the active site for the cleavage but not to catalyze the reaction. Particles that did not cleave in 9 h (∼25%) may not have had the required number of cleavages to stabilize the capsid form and thus reverted to a larger particle not capable of supporting the chemistry when the pH was raised.
The NωV crystal structure shows a large number of acidic residues on adjacent subunit interfaces. At pH 7.6, most of these have a negative charge (Fig. 5) and create electrostatic repulsion that stabilizes the procapsid state. Table 1 shows that many of the acidic residues have calculated pKa values above 5.0 in either the procapsid (based on the pseudoatomic model) or the capsid (based on the X-ray model). These residues gain a proton when the pH nears 5.0 and become neutral, thus reducing the repulsion in the pH range observed for the LCC.
Previously, Taylor et al. mutated a number of residues near the active site of cleavage (25) and showed that assembly was very sensitive to changes in this region. Here we investigated changes to acidic residues to determine their effect on maturation (Table 1). Three mutations from Glu to Gln were made at different residues (Fig. 1), and their cleavage kinetics was examined.
Glu442 has a calculated pKa value above 8.0 in all four subunits of the capsid (Table 1). It is close to the neighboring active site of cleavage (e.g., Glu442 in subunit D approaches the cleavage site of subunit C). The residue was mutated to a Gln to remove the possibility of pH-dependent electrostatic repulsion close to the active site of cleavage (Fig. 5). However, the cleavage rate did not show any significant difference compared with wild-type results (Fig. 6). Thus, both the formation of the active site of cleavage and the chemistry of the cleavage reaction do not depend on this negative charge. In retrospect, the exceptionally high pKa of this residue is consistent with this phenotype, i.e., it is protonated very early in the LCC and therefore behaves almost like the Gln mutation.
Glu73 in subunit B forms a salt bridge with neighboring Arg560 in subunit A (equivalent pairs form between other quasiequivalent subunits). The region around Arg560 includes intramolecular neighbors Asp555 and Asp556, creating a high concentration of negative charge (shown in Fig. 5). It is far from the active site of cleavage (∼39 Å [intramolecular] or ∼26 Å [intermolecular]) and is associated with the intersubunit contacts in the inner helical region of the capsid. To study the effect of the intermolecular interaction involving the helical region, Glu73 was mutated to a Gln to remove the salt bridge. The cleavage for this mutant had an initial rate similar to that seen with the wild type, but the reaction stopped after half the subunits were cleaved (Fig. 6). Since the mutant showed an initial cleavage rate similar to that of the wild type, this substitution was not associated with the pH-dependent LCC required to initiate the cleavage reaction but rather must affect the formation of the cleavage site in half the quasiequivalent subunits. Thus, we propose that rearrangements in the inner helical domain, even distant from the cleavage site, affect cleavage site formation. Further structural study is under way to investigate this mutant.
Glu278 has a calculated pKa value of ∼6.5 in all four subunits of the capsid (Table 1). It is on the polypeptide linking the shell domain and the Ig-like domain and is far from the active site of cleavage. It is widely separated from neighboring subunits in the procapsid and tightly integrated with neighbors near the quasi- and icosahedral threefold axes, penetrating into a cavity that is negatively charged at pH 7.6 but much less repulsive to Glu278 at pH 5.0 (Fig. 5). The residue was mutated to a Gln to remove pH-dependent charge changes (Fig. 5), possibly raising the pH of maturation. In fact, at pH 5.0 virtually no cleavage occurred in the first 30 min and after that a cleavage rate much slower than than that of the wild type was observed. Even at pH 4.5, it was initially slower than the wild-type rate seen at pH 5.0. The cryoEM reconstruction of Glu278Gln was determined at 9.8-Å resolution by flash-freezing particles 2 min after lowering the pH to 5.0, a point where no cleavage had occurred. The spherically averaged outer edge of the capsid density has a radius of 215 Å; Glu278Gln had an outer radius of 235 Å, intermediate between those of the procapsid (255 Å) and capsid (Fig. 8b and 9). The pseudoatomic model of the mutant at pH 5.0 showed that residue 278 did not have the close contact with its neighbor subunit seen in the capsid structure, explaining the intermediate particle size. The negative charge associated with Glu278 interacts with the equivalent residue related to it by threefold or quasithreefold symmetry as the particle changes from procapsid to capsid. The mutual self-repulsion of these symmetry-related residues must be important to guide the transition that allows residue 278 to occupy the cavity in the neighboring subunit (Fig. 8c). Cleavage cannot occur in Glu278Gln at pH 5.0, because the active site for catalysis cannot form with the transition incomplete. Figure 8b and c illustrate the intermediate state for the mutant Gln278. At pH 4.5, approximately 70% of the subunits cleave in 2 h, which is comparable to wild-type results, indicating that the transition is normal at this pH.
The mutagenesis analysis described had proceeded from an assumption that similar phenotypes would be generated in all cases, i.e., that reduction of electrostatic repulsion would allow the LCC and cleavage to occur at a higher pH. In fact, each mutant had its own specific and unexpected phenotype, indicating the complexity of even an apparently straightforward pH-driven transition.
Most animal viruses go though maturation. Picorna- and nodaviruses undergo a subtle maturation that involves autoproteolytic cleavage without an LCC, whereas reovirus undergoes a maturation cleavage, but it has not been possible to characterize a LCC prior to cleavage if, indeed, one exists. Viruses such as human immunodeficiency virus undergo a dramatic change in the morphology of the internal nucleoprotein following proteolysis by the virally encoded protease, and extensive efforts are under way to characterize the mechanism of this LCC (13). The data generated in this study revealed the electrostatic sensitivity that allows NωV to control the time of cleavage and, presumably, infectivity. During the course of infection, tetraviruses induce apoptosis, and this generally lowers the pH of the cell (4). It is likely that the observed maturation occurs late in infection, concurrent with release of the virus particles, at which point they become infectious. Even when the extra cellular environment is at neutral pH, the cleavage continues, following the LCC and cleavage of ∼5% of the subunits.
Acknowledgments
This work was supported by NIH grant R01 GM 54076-12 (to J.E.J.). Electron microscopic imaging and reconstruction were conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through National Center for Research Resources program P41 (grant RR17573).
We thank Teddy Ajero for technical help with the electron microscopy and Nathan Baker at Washington University, St. Louis, MO, for help in calculation of the electrostatic potentials with his program ProPKA and for helpful discussions. We appreciate the constructive criticism provided by Jeffrey Speir and Kelly Lee.
Footnotes
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Agrawal, D. K., and J. E. Johnson. 1995. Assembly of the T = 4 Nudaurelia capensis omega virus capsid protein, post-translational cleavage, and specific encapsidation of its mRNA in a baculovirus expression system. Virology 20789-97. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, D. K., and J. E. Johnson. 1992. Sequence and analysis of the capsid protein of Nudaurelia capensis omega virus, an insect virus with T = 4 icosahedral symmetry. Virology 190806-814. [DOI] [PubMed] [Google Scholar]
- 3.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 9810037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, E. M., K. H. Gordon, S. J. Dorrian, E. R. Hines, and T. N. Hanzlik. 2002. Infection of its lepidopteran host by the Helicoverpa armigera stunt virus (Tetraviridae). J. Invertebr. Pathol. 8097-111. [DOI] [PubMed] [Google Scholar]
- 5.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54905-921. [DOI] [PubMed] [Google Scholar]
- 6.Canady, M. A., M. Tihova, T. N. Hanzlik, J. E. Johnson, and M. Yeager. 2000. Large conformational changes in the maturation of a simple RNA virus, nudaurelia capensis omega virus (NomegaV). J. Mol. Biol. 299573-584. [DOI] [PubMed] [Google Scholar]
- 7.Canady, M. A., H. Tsuruta, and J. E. Johnson. 2001. Analysis of rapid, large-scale protein quaternary structural changes: time-resolved X-ray solution scattering of Nudaurelia capensis omega virus (NomegaV) maturation. J. Mol. Biol. 311803-814. [DOI] [PubMed] [Google Scholar]
- 8.Carragher, B., N. Kisseberth, D. Kriegman, R. A. Milligan, C. S. Potter, J. Pulokas, and A. Reilein. 2000. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 13233-45. [DOI] [PubMed] [Google Scholar]
- 9.Caspar, D. L., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 271-24. [DOI] [PubMed] [Google Scholar]
- 10.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50760-763. [DOI] [PubMed] [Google Scholar]
- 11.Dolinsky, T. J., P. Czodrowski, H. Li, J. E. Nielsen, J. H. Jensen, G. Klebe, and N. A. Baker. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35W522-W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolinsky, T. J., J. E. Nielsen, J. A. McCammon, and N. A. Baker. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32W665-W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganser-Pornillos, B. K., M. Yeager, and W. I. Sundquist. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18203-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanzlik, T. N., S. J. Dorrian, K. H. Gordon, and P. D. Christian. 1993. A novel small RNA virus isolated from the cotton bollworm, Helicoverpa armigera. J. Gen. Virol. 74(Pt. 9)1805-1810. [DOI] [PubMed] [Google Scholar]
- 15.Hanzlik, T. N., and K. H. Gordon. 1997. The Tetraviridae. Adv. Virus Res. 48101-168. [PubMed] [Google Scholar]
- 16.Helgstrand, C., S. Munshi, J. E. Johnson, and L. Liljas. 2004. The refined structure of Nudaurelia capensis omega virus reveals control elements for a T = 4 capsid maturation. Virology 318192-203. [DOI] [PubMed] [Google Scholar]
- 17.Li, H., A. D. Robertson, and J. H. Jensen. 2005. Very fast empirical prediction and rationalization of protein pKa values. Proteins 61704-721. [DOI] [PubMed] [Google Scholar]
- 18.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 12882-97. [DOI] [PubMed] [Google Scholar]
- 19.Mallick, S. P., B. Carragher, C. S. Potter, and D. J. Kriegman. 2005. ACE: automated CTF estimation. Ultramicroscopy 1048-29. [DOI] [PubMed] [Google Scholar]
- 20.Munshi, S., L. Liljas, J. Cavarelli, W. Bomu, B. McKinney, V. Reddy, and J. E. Johnson. 1996. The 2.8 A structure of a T = 4 animal virus and its implications for membrane translocation of RNA. J. Mol. Biol. 2611-10. [DOI] [PubMed] [Google Scholar]
- 21.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 22.Shepherd, C. M., I. A. Borelli, G. Lander, P. Natarajan, V. Siddavanahalli, C. Bajaj, J. E. Johnson, C. L. Brooks III, and V. S. Reddy. 2006. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 34D386-D389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speir, J., and J. Johnson. 2008. Tetravirus structure, p. 27-37. Encyclopedia of virology, 3rd ed., vol. 5. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 24.Steven, A. C., J. B. Heymann, N. Cheng, B. L. Trus, and J. F. Conway. 2005. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr. Opin. Struct. Biol. 15227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor, D. J., and J. E. Johnson. 2005. Folding and particle assembly are disrupted by single-point mutations near the autocatalytic cleavage site of Nudaurelia capensis omega virus capsid protein. Protein Sci. 14401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor, D. J., N. K. Krishna, M. A. Canady, A. Schneemann, and J. E. Johnson. 2002. Large-scale, pH-dependent, quaternary structure changes in an RNA virus capsid are reversible in the absence of subunit autoproteolysis. J. Virol. 769972-9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu, Y., B. Carragher, F. Mouche, and C. S. Potter. 2003. Automatic particle detection through efficient Hough transforms. IEEE Trans. Med. Imaging 221053-1062. [DOI] [PubMed] [Google Scholar]