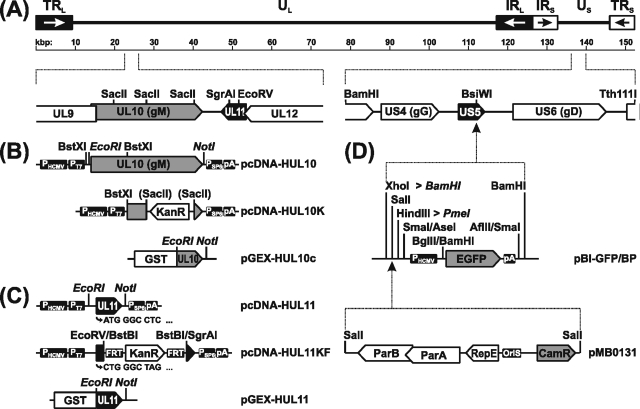
FIG. 1.
Construction of expression plasmids and virus mutants. (A) A schematic map of the HSV-1 genome shows the unique long (UL) and unique short (US) regions flanked by terminal (TRL and TRS) and internal inverted (IRL and IRS) repeat sequences. Numbers denote kilobase pairs. Relevant ORFs (pointed rectangles) and restriction sites are indicated. (B) Eukaryotic expression plasmid pcDNA-HUL10 contains the gM gene flanked by the HCMV immediate-early promoter (PHCMV) and a polyadenylation signal (pA). In pcDNA-HUL10K part of UL10 was replaced by a kanamycin resistance gene (KanR), and the PCR product obtained with PT7- and PSP6-specific primers was used for construction of gM deletion mutants. For preparation of a monospecific rabbit antiserum, the C terminus of gM was expressed as a bacterial fusion protein with GST from pGEX-HUL10. (C) Corresponding prokaryotic and eukaryotic expression plasmids were also generated for the complete UL11 gene (pGEX-HUL11 and pcDNA-HUL11). Deletion plasmid pcDNA-HUL11KF permitted removal of the KanR gene using flanking FRTs and introduction of a mutated start codon (CTG) as well as an artificial stop codon (TAG) to prevent expression of the 5′ part of UL11. (D) The genome of HSV-1 strain KOS was cloned as a BAC after insertion of a mini-F plasmid vector (pMBO131), together with an expression cassette for EGFP at the nonessential US5 gene encoding gJ. Artificial BamHI and PmeI restriction sites (shown in italic) were created to facilitate cloning and mutagenesis.