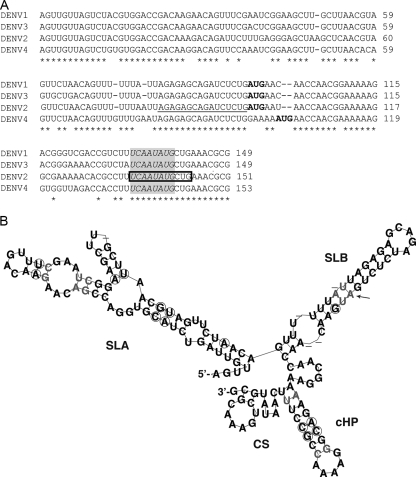
FIG. 1.
Predicted and solution structures of the 5′ end of DENV2. (A) CLUSTAL W (v.1.83) alignment of the first ∼150 nt of DENV1 to DENV4. The CS region is boxed, and conserved nucleotides among the flaviviruses (dengue, yellow fever, Murray Valley encephalitis, West Nile, and St. Louis encephalitis viruses) (6) are in italics in the shaded area. The start codon is indicated in boldface, and the UAR is underlined. Asterisks indicate conserved nucleotides among all DENV serotypes. (B) Consensus structure prediction of DENV serotypes from panel A, generated by Alifold. Circles indicate sequence variation supporting the consensus structure. Pairs with noncompatible sequence variation are shown in gray, and dashes represent variable positions. The arrow indicates the start codon.