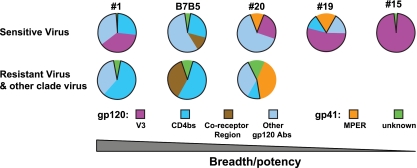
FIG. 5.
Schematic summary of serum neutralization specificities. The data are shown as pie charts that summarize the fraction of the neutralization activity defined in selected sera. Both the gp120 and gp41 components and subspecificities within these two elements of Env are shown as averages for the two virus types (see below), and the values are derived from Tables 1, 2, 3, and 4. Significant neutralizing specificity pattern differences of the HIV-positive sera against sensitive viruses (top pie charts, HXBc2, SF162, BaL.01, and JRFL-Δ301) and resistant viruses (JRFL, PVO.04 and other clade viruses) are shown. For sensitive viruses, the narrowly neutralizing sera show a predominant V3-directed neutralizing specificity, while the broadly neutralizing sera display lower levels of V3-directed neutralizing specificity. Resistant viruses are neutralized only by the broadly neutralizing and potent sera. Note that most neutralizing activity maps to gp120, and several samples, including B7B5, exhibit CD4bs-directed activity. Serum B7B5 also contains neutralizing activity that is affected by the coreceptor binding site mutation. Sera 19 and 20 contain detectable MPER-directed neutralization, but of limited potency. Abs, antibodies.