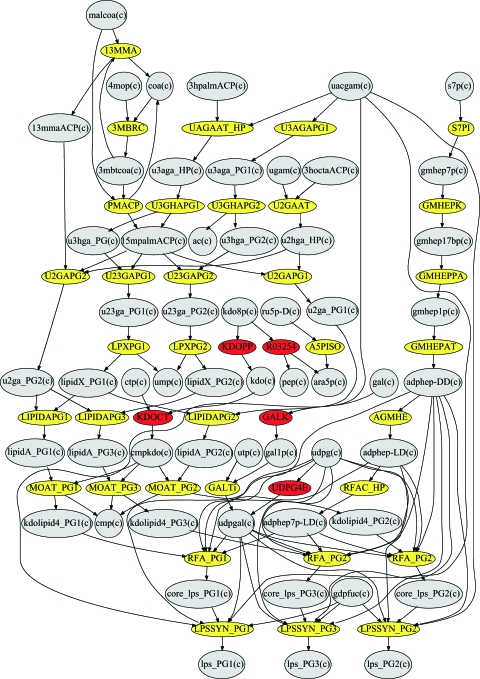
FIG. 7.
Representation of the putative pathways for LPS biosynthesis in the iVM679 P. gingivalis model, with layout automatically generated by using GraphViz (see Materials and Methods). Gray nodes denote metabolites; red and yellow nodes represent reactions. The red nodes denote reactions that have been linked to specific reactions. Yellow nodes represent reactions that are putative and that were manually added in order to make all three specific lipid A molecules. The UDPG4E reaction is linked to the galE gene, producing a UDP-glucose 4-epimerase. The galE mutants are known to have modified lipid A molecules (59). The pathway culminates in the production of lps_PG1, lps_PG2, and lps_PG3 molecules, which refer to the PG1690, PG1450, and PG1435 lipid A structures, respectively.