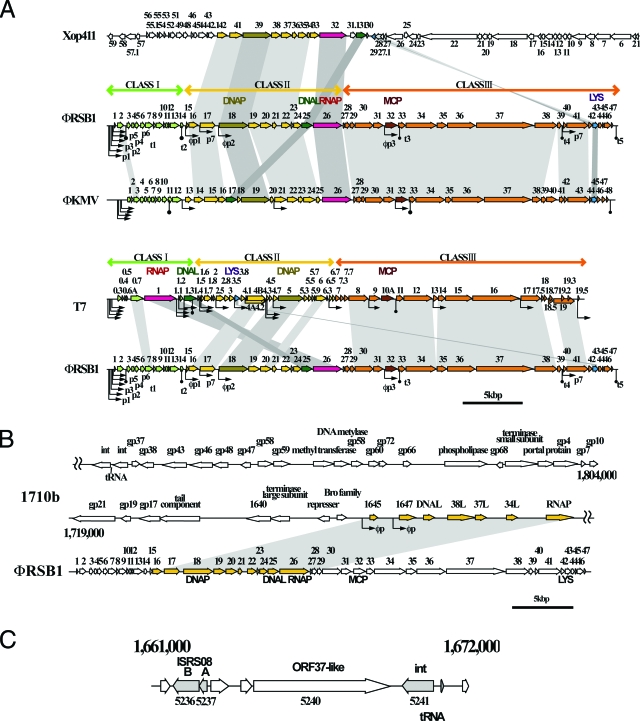
FIG. 1.
Genetic organization and comparative analyses of the φRSB1 genome. (A) Comparison of the genomes of φRSB1 and T7-like phages. In each alignment, corresponding ORFs (horizontal arrows) are connected by shading. Three functional gene clusters, class I (green), class II (yellow), and class III (orange), are indicated above the φRSB1 and the T7 maps, and corresponding ORFs are colored. Putative bacterial promoters, phage promoters, and terminators of transcription are indicated under the φRSB1 map by p, φp, and t, respectively. Promoters and terminators are also shown in the φKMV and T7 maps. Xop411, Xanthomonas oryzae phage (44,520 bp, accession no. DQ777876); φKMV, Pseudomonas aeruginosa phage (42,519 bp, AJ50558); T7, coliphage T7 (39,937 bp, NC_00164). DNAP, DNA polymerase; MCP, major capsid protein; LYS, lysozyme. (B) The class II region of the φRSB1 genome (ORF16 to ORF26) often shows high homology with phage or prophage sequences of different phage groups. The φRSB1 region is aligned with the corresponding prophage sequence of Burkholderia pseudomallei 1710b (accession no. CP000124). (C) Region on the GMI1000 genome (positions 1661000 to 1672000) containing a large φRSB1 ORF37-like ORF (RSO5240), which encodes a putative transglycosylase protein. This GMI1000 region is flanked by two ORFs encoding ISRS08 transposase A and B on the left side and by integrase (RS05241) and arginine tRNA (AGA) on the right side.