Abstract
Prebiotics such as fructooligosaccharides (FOS) are increasingly being used in some countries for improving human and animal health and as an alternative to antibiotic growth promoters in animals, with various degrees of success. It has been observed that FOS stimulate the proliferation of probiotic bacteria and, at the same time, decrease the population of bacteria associated with disease. This observation assumes that pathogenic bacteria do not metabolize FOS and, therefore, lose their competitive advantage over beneficial bacteria. Here we present evidence that some pathogenic Escherichia coli strains can metabolize FOS and show that this property helps the bacterium colonize the intestine. These findings highlight the potential risk that a high level of prebiotic usage could lead to the emergence of well-adapted pathogenic strains that metabolize prebiotic substances.
A prebiotic is a selectively fermented ingredient which allows specific changes in the composition of and/or activity in the gastrointestinal microbiota which confer health benefits on the host (37). Many carbohydrates, including short-chain fructooligosaccharides (scFOS), are reported to be prebiotic. scFOS are polymers of fructose units with the generic structure α-d-Glu-(1-2)-(β-d-Fru-1-2-)n, where n is between 2 and 4. FOS are low-energy ingredients with a taste similar to that of sucrose. They are used commercially in food products and nutritional supplements. In the intestines of humans, rats, horses, pigs, and chicks, FOS stimulate growth of probiotic bacteria, and some studies have found that they inhibit the growth of pathogenic bacteria such as Salmonella enterica serovar Typhimurium and Escherichia coli (4, 7, 33, 42). Genes involved in the assimilation of FOS by probiotic bacteria, such as lactic acid bacteria, have been identified (3, 21, 39), but so far they have not been found in pathogenic bacteria.
Extraintestinal pathogenic E. coli strains are normal inhabitants of the guts of humans and warm-blooded animals. They are the major cause of extraintestinal infections, being the principal agent of urinary tract infections, the second most common agent of neonatal meningitis, and one of the two most common agents of bacteremia (25). They are also responsible for intra-abdominal, soft tissue, and respiratory tract infections (38). The latter infection is particularly found in poultry and often leads to air sacculitis, perihepatitis, and pericarditis, as well as other syndromes, such as osteomyelitis (2, 16).
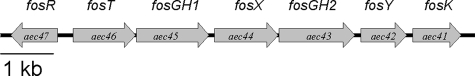
In order to colonize different niches, E. coli has acquired new functions by horizontal gene transfer (29). Much of this horizontally transferred DNA is clustered into genomic islands (17). We previously identified a pathogenicity island, named AGI-3 (GenBank accession number AY857617), in the chromosome of an extraintestinal avian-pathogenic strain of E. coli, BEN2908. AGI-3 shows a modular structure composed of five loci bound by mobility-related genes (10). Three genes of the first locus are related to carbon metabolism and are involved in the virulence of the strain for chickens at an early stage of infection. The second locus (termed locus 2), comprising genes aec41 to aec47, is also thought to be involved in the assimilation of carbohydrates. Homology searches indicate that it codes for a transcriptional regulator of the LacI family (Aec47), a sugar transporter of the major facilitator superfamily (Aec46), two proteins of unknown functions (Aec42 and Aec44), a fructokinase (Aec41), and two glycoside hydrolases (Aec43 and Aec45) (Fig. 1) (10). Glycoside hydrolases (EC 3.2.1.) are a widespread group of enzymes that hydrolyze the glycosidic bond between two or more carbohydrates or between a carbohydrate and a noncarbohydrate moiety. They have been grouped into families based on their overall amino acid sequence similarities (12). The two glycoside hydrolases encoded by locus 2 genes belong to family 32 in the carbohydrate-active enzyme database (http://www.cazy.org). Enzymes of this family are functionally related and are involved in the hydrolysis of glycosidic bonds of fructose polymers, such as inulin and fructans. To be metabolized by E. coli, fructose has to be phosphorylated (18). E. coli does not usually possess cytoplasmic fructokinase. Fructose enters the cell via a phosphotransferase system (PTS), and phosphorylation is concomitant with membrane transport (28). The fact that locus 2 putatively encodes a fructokinase and glycoside hydrolases hydrolyzing fructose-containing polymers suggests that the carbohydrate transported and metabolized via locus 2 contains fructose units.
FIG. 1.
Gene organization of the fos locus of E. coli strain BEN2908. The locus comprises fosR (aec47, putative regulator), fosT (aec46, putative sugar transporter), fosGH1 (aec45, putative glycoside hydrolase), fosX (aec44, unknown function), fosGH2 (aec43, putative glycoside hydrolase), fosY (aec42, unknown function), and fosK (aec41, putative fructose kinase).
This study shows that locus 2 of the pathogenicity island AGI-3 is involved in scFOS metabolism. By comparing the abilities of the wild-type strain BEN2908 and its isogenic mutant, which is unable to metabolize scFOS, to colonize the intestine, we demonstrate that locus 2 contributes to the intestinal fitness of the pathogenic E. coli strain BEN2908.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The main bacterial strains and plasmids used in this study are listed in Table 1. E. coli strain BEN2908, O2:K1:H5 (Fim+ Iut+ IbeA+ AGI-3+), is a nalidixic acid-resistant derivative of strain MT78 which was isolated from the trachea of a chicken with a respiratory infection (10, 15, 20). E. coli strain BEN2908 belongs to the phylogenetic group B2 (32). For cloning experiments, E. coli strains XL1-Blue and MG1655 were used (5, 8). For phenotypic and genotypic studies, a total of 133 strains were used; they included 72 strains from the ECOR collection (E. coli reference collection containing 61 fecal isolates from humans and animals, 10 urinary tract infection isolates, and 1 asymptomatic bacteriuria isolate from humans), 34 strains from humans with extraintestinal syndromes (meningitis or septicemia), and 27 strains from chickens that tested positive for the first locus of AGI-3 (4 nonpathogenic and 23 extraintestinal pathogenic strains) (10, 34). Strains were routinely grown in Luria-Bertani (LB) medium at 37°C with agitation. When necessary, kanamycin or ampicillin was added at a final concentration of 50 μg/ml or 100 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| E. coli strains | ||
| BEN2908 | Extraintestinal pathogenic strain; O2:K1:H5 (Nalr Fos+ Fim+ Iut+ IbeA+ AGI-3+), avian origin | 15 |
| BEN2908ΔfosT | Isogenic deletion mutant of BEN2908; Kanr Fos− | This study |
| MG1655 | Nonpathogenic, Fos−, human origin | 5 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pKD4 | oriRγ Ampr Kanr | 14 |
| pKD46 | oriR101 repA101(ts) araBp-gam-bet-exo Ampr | 14 |
| pGEM-T easy vector | Ampr | Promega |
| pGEM::fos | pGEM containing the whole fos locus | This study |
For cloning experiments, plasmid pGEM-T easy vector was used (Promega).
Construction of the fosT mutant and cloning of the fos locus.
The fosT knockout mutant was constructed by using the method developed by Datsenko and Wanner and as applied previously (10, 14). In brief, after recombination, fosT was deleted and replaced by a kanamycin resistance cassette obtained by PCR amplification of plasmid pKD4 by using primers cat147 and cat148, which contain extensions homologous to the 5′ and 3′ ends of fosT (see Table S1 in the supplemental material). The replacement of fosT was confirmed by PCR using the primer pairs px1/cat162 and px2/cat156 (see Table S1 in the supplemental material), which made it possible to detect the left and right arms of the insertion, respectively. To clone the fos locus, the whole locus (9,027 bp) was amplified from strain BEN2908 by PCR using the primer pair cat154/cat155 (see Table S1 in the supplemental material) and Herculase enhanced DNA polymerase (Stratagene). The resulting PCR product was inserted into the PGEM-T easy vector (Promega). The ligation product was electroporated into competent XL1-Blue, prepared as described by Tung and Chow (44). Recombinant plasmids were analyzed by restriction analysis using PstI and BamHI (Promega) and by PCR using primer pairs cat167/cat168 and cat163/cat164 (see Table S1 in the supplemental material), allowing the amplification of the junctions between fosY and fosK and between fosGH2 and fosX, respectively. A recombinant pGEM::fos plasmid was then introduced by electroporation into electrocompetent MG1655 cells.
Growth monitoring.
For growth monitoring experiments, overnight LB cultures were centrifuged, washed twice, and resuspended in the same volume of M9 minimal medium (31). The strains were then cultured in triplicate at 37°C in 100-well, sterile, covered microplates (Labsystems, Helsinki, Finland). Each well contained 300 μl of M9 medium supplemented with either 0.5% scFOS (Profeed P95; Beghin Meiji, France), 0.2% glucose (Sigma), 5 mM kestose (GF2; Wako Chemicals GmbH, Germany), 5 mM nystose (GF3; Wako Chemicals GmbH, Germany), or 5 mM fructofuranosyl nystose (GF4; Wako Chemicals GmbH, Germany). The plates were incubated in a Microbiology Reader Bioscreen C apparatus (Labsystems, Helsinki, Finland), and the turbidity from 405 to 600 nm was measured every 30 min, after shaking. For analysis of the FOS+ phenotype, overnight LB cultures were centrifuged, washed twice, and resuspended in the same volume in M9 minimal medium. The strains were then cultured overnight in 5 ml of M9 medium supplemented with 0.5% scFOS or 0.2% glucose. A negative control experiment was performed using M9 medium with no carbon source.
Detection of fos genes in E. coli isolates.
The presence of fos genes in E. coli isolates was analyzed by PCR using primer pairs Rorf10/Forf10 and cat49/cat50, allowing the detection of the fosGH2 gene and the fosR-fosT intergenic region, respectively (see Table S1 in the supplemental material). PCRs were performed using a 25-μl volume containing 500 nM of the forward and reverse primers, 200 μM of each deoxynucleoside triphosphate (Finnzymes, Ozyme, France), 1 U of Taq DNA polymerase (New England Biolabs Inc.), and 2 mM MgCl2 in a PCR buffer containing 1 mM KCl, 1 mM (NH4)2SO4, 200 μM MgSO4, 0.1% Triton X-100, 2 mM Tris-HCl (pH 8.8) (New England Biolabs Inc.), and 5 μl of DNA template prepared by the boiling method (40). The PCR conditions were as follows: initial denaturation at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1 min/kb. A final extension step at 72°C for 7 min was included. Reactions were performed in a PerkinElmer thermocycler (GeneAmp 9700; Applied Biosystems).
Intestinal colonization of chickens.
Axenic strain PA12 White Leghorn chicks were obtained from the INRA infectiology platform by using the method described by Le Bars (30). Specific-pathogen-free (SPF) chickens were obtained by orally inoculating 8-day-old axenic chicks with a complex bacterial inoculum consisting of 0.5 ml of a 1/10 suspension of feces from adult SPF hens (6). The E. coli strains in this inoculum were all antibiotic sensitive. The housing, husbandry, and slaughtering conditions conformed to European Union guidelines for the care and use of laboratory animals. The experimental protocol was approved by the regional ethical committee under number CL2007-43. Animals were reared in isolators, fed ad libitum on a commercial diet sterilized by gamma irradiation (poultry starter HPS; Dietex, France), and supplied with autoclaved tap water or 0.5% autoclaved scFOS throughout the experiment. After 18 h of food starvation, eight 12-day-old axenic chicks or eight 12-day-old SPF chicks were fed with 0.5 ml of a mixture of equal numbers of bacteria of the wild-type strain BEN2908 Nalr and the mutant derivative BEN2908ΔfosT Kanr Nalr (each approximately at 5 × 106 CFU) in LB medium. Fresh fecal samples were collected on days 1, 2, 3, 6, 8, 10 or 13, 15, and 20 or 21 postgavage. Fecal samples were weighed and then homogenized in distilled water (9 ml/g of feces). Viable E. coli cells were counted by plating 10-fold dilutions in sterile saline on Drigalski agar (Bio-Rad), with nalidixic acid at 40 μg/ml (selections of the wild-type and ΔfosT strains) or with kanamycin at 50 μg/ml (selection of the ΔfosT strain). The numbers of fecal CFU were calculated per gram of feces. The detection threshold was 100 CFU/g of feces. The numbers of wild-type bacterial CFU (BEN2908 Nalr) were calculated by subtracting the number of kanamycin-resistant bacteria from the number of nalidixic-resistant bacteria. Competition indices (CI) were calculated similarly to the method of Freter et al. by using BEN2908 as the reference strain [CI = (number of BEN2908 CFU/number of BEN2908ΔfosT CFU recovered from chicken feces)/(number of BEN2908 CFU/number of BEN2908ΔfosT CFU present in the initial inoculum)] (19). By definition, a CI of >1 indicates outcompetition of the mutant strain (BEN2908ΔfosT) by the wild-type reference strain (BEN2908). A CI equal to 1 indicates no difference in colonization, and a CI of <1 indicates outcompetition of the wild-type reference strain (BEN2908) by the mutant (BEN2908ΔfosT). In all colonization experiments, we verified that the kanamycin-resistant E. coli population was unable to metabolize scFOS. To that end, 5 ml of LB medium containing kanamycin at 50 μg/ml was inoculated with 100 μl of each fecal sample and incubated overnight at 37°C with agitation. Overnight LB cultures were centrifuged, washed twice with M9 minimal medium, and resuspended in the same volume of M9 minimal medium. Five milliliters of M9 minimal medium supplemented with 0.5% scFOS was then inoculated with 100 μl of the washed culture and incubated overnight at 37°C with agitation. No growth was observed for any of the fecal samples tested.
Statistical analysis.
Statistical analyses of CI were performed by using the Mann-Whitney U test. Exact P values were calculated with StatXact software (version 5.0; Cytel Inc., Cambridge, MA).
RESULTS
Involvement of locus 2 of the pathogenicity island AGI-3 in the metabolism of FOS.
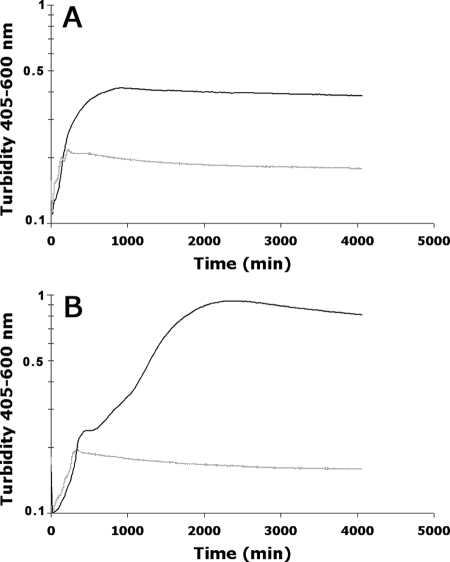
Since in silico analysis of locus 2 of the pathogenicity island AGI-3 of BEN2908 suggested that it is involved in the metabolism of a sugar containing fructose units, we tested the ability of BEN2908 to grow in minimal medium containing different fructose polymers as the sole carbon source. We found that BEN2908 was not able to grow in minimal medium containing sucrose, raffinose, or inulin but thrived in the presence of scFOS (data not shown and Fig. 2A). This indicates that locus 2 of AGI-3 could play a role in the metabolism of scFOS.
FIG. 2.
The metabolism of scFOS is mediated by locus 2 of AGI-3. E. coli strains BEN2908 (solid black curve) and BEN2908Δaec46 (dashed gray curve) (A) or MG1655 (dashed gray curve) and MG1655/pGEMT::aec41-47 (solid black curve) (B) were grown in M9 medium containing 0.5% scFOS at 37°C with agitation.
To prove that locus 2 is involved in scFOS metabolism, we first constructed a derivative of strain BEN2908 by replacing the aec46 gene, encoding the putative scFOS transporter, with a kanamycin resistance cassette. We then tested the ability of the mutant strain to grow in a minimal medium with scFOS as the sole carbon source. Figure 2A indicates that BEN2908Δaec46 is unable to grow in this medium. Secondly, we introduced a recombinant plasmid carrying the whole locus 2 in the nonpathogenic E. coli strain MG1655, which is unable to metabolize scFOS. The recombinant MG1655 strain was now able to grow with scFOS as the sole carbon source (Fig. 2B). Taken together, these two experiments prove that the second locus of AGI-3 is involved in scFOS metabolism. Accordingly, we renamed locus 2 the fos locus and the individual genes of locus 2 fosR (aec47, putative regulator), fosT (aec46, scFOS transporter), fosGH1 (aec45, putative glycoside hydrolase), fosX (aec44, unknown function), fosGH2 (aec43, putative glycoside hydrolase), fosY (aec42, unknown function), and fosK (aec41, putative fructose kinase) (Fig. 1).
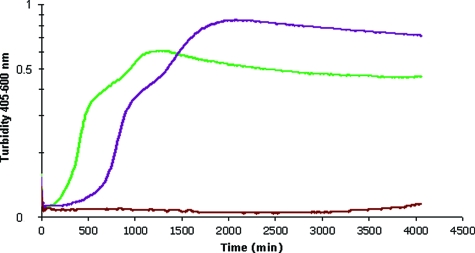
Different sugar polymers belong to the family of scFOS, depending on their numbers of fructose units. To identify which type of scFOS is consumed by strain BEN2908, we monitored the growth of BEN2908 in M9 minimal medium supplemented with either 5 mM of kestose (2 units of fructose [GF2]), nystose (3 units of fructose [GF3]), or fructofuranosylnystose (4 units of fructose [GF4]). It was found that BEN2908 metabolized GF2 and GF3 at the same rate but consumed GF2 earlier than GF3. GF4 did not allow the growth of BEN2908 (Fig. 3).
FIG. 3.
Preferential use of the shorter-chain FOS. E. coli strain BEN2908 was grown in M9 medium containing either 5 mM GF2 (green curve), 5 mM GF3 (purple curve), or 5 mM GF4 (red curve) at 37°C with agitation.
Effect of the fos locus on intestinal colonization.
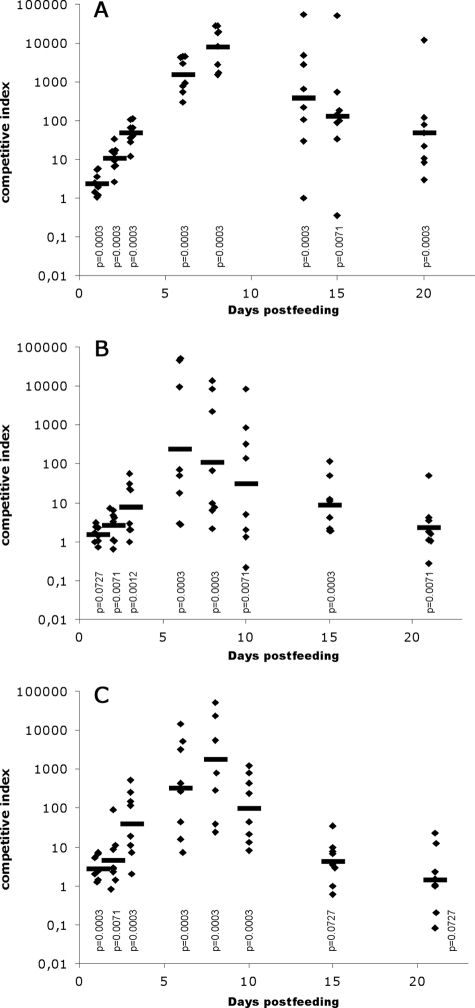
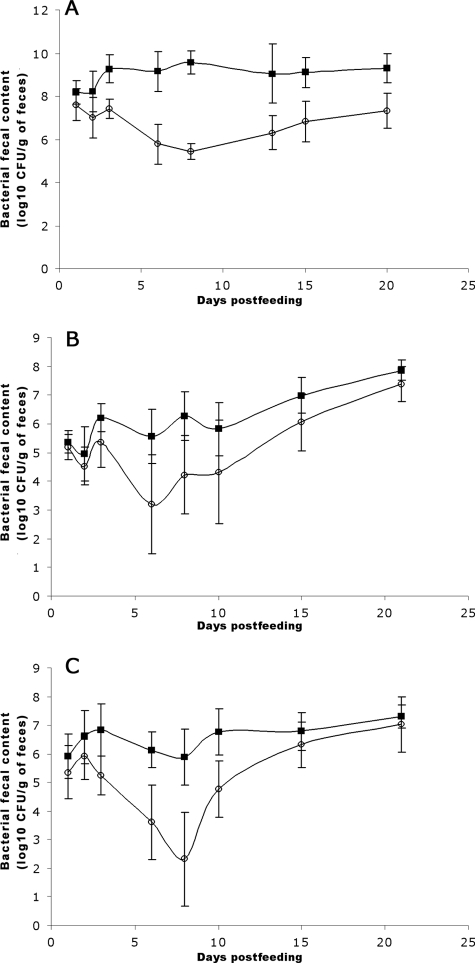
FOS are naturally found in many plants, such as wheat (13), and consequently are present in chicken feed and in the intestine, the primary habitat of extraintestinal pathogenic E. coli. We thus examined the effect of the fos locus on the ability of E. coli BEN2908 to colonize the chicken intestine. To that end, E. coli BEN2908ΔfosT (formerly BEN2908Δaec46) and its wild-type parent were fed together in equal amounts to axenic chickens (5 × 106 CFU of each strain/chicken), and the proportion of each in feces was monitored. Figure 4A indicates that the mutant strain was strongly outcompeted by the wild-type strain. The mean CI increased up to day 8 postfeeding (mean CI of 8,078) and then decreased, although it was still substantial at day 20 (mean CI of 48). This decrease was not a consequence of a global E. coli population decrease due to the development of mucosal immunity, since more than 109 CFU/g of feces were still present at day 20 (Fig. 5A). These results indicate that the fos locus is beneficial during the initial stage of colonization and less important during the maintenance stage.
FIG. 4.
Competition between E. coli strains BEN2908 and BEN2908ΔfosT to colonize the chicken intestine. Axenic chickens (A), SPF chickens (B), or SPF chickens receiving a dietary supplement of 0.5% scFOS in drinking water (C) were fed with strains BEN2908 and BEN2908ΔfosT together (each at 5 × 106 CFU/chicken). The proportion of each strain in animal feces was monitored over time, and CI were calculated. Horizontal bars indicate the geometric means of CI, and diamonds indicate individual CI. Statistical analyses were conducted using the Mann-Whitney U test, measuring the difference between CI in feces samples and in the inoculum. The calculated P values are presented, with values below 0.05 considered significant.
FIG. 5.
Monitoring of chicken intestine colonization by E. coli strain BEN2908 (filled squares) and its mutant derivative BEN2908ΔfosT (open circles). Axenic chickens (A), SPF chickens (B), or SPF chickens receiving a dietary supplement of 0.5% scFOS in drinking water (C) were fed with strains BEN2908 and BEN2908ΔfosT together (each at 5 × 106 CFU/chicken). The presence of each strain in the feces was monitored over time. Bars represent standard errors of the log10 (mean number of CFU/g of feces) for each set of eight chickens.
To mimic more natural conditions, we repeated this experiment with SPF chickens which have a complex biota, containing commensal E. coli among others bacteria. Even when this complex biota is found in the intestine, our findings indicate that strain BEN2908 is perfectly able to colonize the intestine (108 CFU/g feces at day 21 postfeeding) (Fig. 5B) and to outcompete the ΔfosT mutant, reaching lower but still highly competitive levels (Fig. 4B). This indicates that scFOS metabolism provides selective advantages for strain BEN2908 within the highly competitive gastrointestinal tract at the expense of a similar strain that does not metabolize it. The fact that mean CI were lower in the presence of a complex microbiota than in axenic chickens may suggest that other bacteria of the flora are more efficient FOS degraders than strain BEN2908.
Dietary supplements of FOS have been used at poultry farms to improve the health and performance of poultry (36). We therefore also evaluated the impact of increasing the scFOS content of the chicken diet (0.5% in drinking water) on the capacity of strains BEN2908 and BEN2908ΔfosT to colonize the intestines of SPF chickens (Fig. 4C). Although the mean CI was lower at day 8, when the chicken diet was not supplemented with scFOS, than when it was supplemented with 0.5% scFOS, this difference was not found to be statistically significant (Fig. 4B and C). This result indicates that dietary supplementation of scFOS does not increase the ability of strain BEN2908 to colonize the intestine.
Other E. coli strains are able to metabolize scFOS.
To determine whether the metabolism of scFOS is a property unique to BEN2908, we performed a rapid survey of various E. coli strains, testing the 72 strains of the E. coli reference collection (ECOR), 27 E. coli strains of avian origin that previously tested positive for the first locus of AGI-3, and 34 strains isolated from humans with extraintestinal syndromes (10, 34). Phenotypic analysis indicated that 4 of the 72 strains of the ECOR collection are able to metabolize FOS; these 4 strains were all isolated from healthy animals (ECOR7 from an orangutan, ECOR23 from an elephant, ECOR31 from a leopard, and ECOR32 from a giraffe). Only one of these strains, ECOR31, was positive when the presence of fosGH2 and overlapping fosT and fosR fragments were tested using PCR. Among the 27 strains of avian origin possessing the first locus of AGI-3 (24 extraintestinal pathogenic strains and 3 nonpathogenic strains), 17 were able to grow with scFOS as the sole carbon source and 9 were PCR test positive (1 of these 9 strains is nonpathogenic). Of the 34 strains isolated from humans with extraintestinal syndromes (meningitis or septicemia), 2 were able to grow in the presence of scFOS but were PCR test negative. Thus, some commensal and pathogenic E. coli strains also possess the ability to metabolize scFOS via the fos locus described here. However, other genes which have not yet been identified are probably implicated in scFOS metabolism in E. coli.
DISCUSSION
FOS is used as a food additive in Europe and Japan for its bifidogenic effect on human colonic endogenous biota, among other things. FOS is considered a protective nutrient, leading to a decrease in the pathogenic population. It has been suggested that probiotic bacteria in the intestinal tract, such as lactobacilli and bifidobacterial species, preferentially use FOS (26). There is increasing evidence that lactobacilli and bifidobacteria develop antimicrobial activities which are involved in the host's gastrointestinal defense system (41). These antimicrobial activities are due partly to the production of lactic acid and an increase in short-chain fatty acid production, resulting in a lower pH in the large intestine which could suppress or displace undesirable or pathogenic bacteria, such as Clostridium perfringens, Salmonella enterica serovar Typhimurium, and E. coli (1, 33, 43). This study demonstrates that, like many probiotic bacteria of the biota, some pathogenic E. coli strains are able to metabolize FOS. This metabolism of FOS helps the strain colonize the intestine, especially in the initial stages. This growth advantage for a pathogenic E. coli strain could suppress the beneficial effect provided by the ingestion of FOS.
E. coli is a very versatile bacterium with high genetic diversity. The plasticity of its genome is mainly the result of gene rearrangement within the genome and of the acquisition of novel traits by horizontal transfer (plasmids, bacteriophages, transposons, or genomic islands). The FOS metabolic genes identified in this study are located in the pathogenicity island AGI-3, whose characteristics are linked to the mobility of the island (presence of direct repeats flanking the island and of an integrase gene) (10, 24). This suggests that AGI-3 has the capacity to be mobile, a property which could contribute to the emergence of the FOS metabolic trait among pathogenic E. coli strains and other pathogenic bacteria of the intestinal biota under selective pressure for FOS utilization. The dissemination of the fos locus would lead to a global increase in the intestinal fitness of the recipient strains and in the risk to public health, thus negating the expected positive effects provided by the use of FOS.
The fos locus presented here has never been reported before, and its origin is still unknown. Various genes involved in FOS metabolism have previously been described to occur in lactic acid bacteria. A gene cluster encoding an ATP-dependent binding cassette (ABC)-type transporter involved in the uptake of scFOS has been reported in Lactobacillus acidophilus (3). Lactobacillus paracasei transports scFOS via an ABC transport system or a PTS (21, 26). FOS uptake in Bifidobacterium longum may be mediated via an ABC-type transporter (23, 35). All of these systems are different from the fos locus of strain BEN2908. The homolog closest (54% identity) to the BEN2908 FosT transporter is a non-PTS sugar permease called FruP which has been identified in Bacillus megaterium but for which the transported substrate has not been identified (9). The homologs closest to the glycoside hydrolases FosGH1 and FosGH2 are a β-fructofuranosidase from Bifidobacterium breve (41% identity) and a glycoside hydrolase from Bacteroides thetaiotaomicron (28% identity), respectively (39, 46).
The digestive tracts of herbivores differ structurally from those of carnivores with regard to the site of cellulose digestion. The large intestine of birds consists of a shorter colon than that of mammals and typically have a pair of ceca. This leads to less efficient recovery of the nutrients from feedstuffs (45). Despite these anatomical and physiological differences between birds and mammals, the composition of the microbiota from the gastrointestinal tracts of chickens is largely similar to the human complex microbial ecosystem, although individual differences in the human microbiota have been found (11, 22, 27). Furthermore, numerous studies have shown that E. coli strains (and particularly strain BEN2908 used in this study) isolated from chickens and humans are genetically similar (32). Our findings suggest that also in mammals pathogenic E. coli strains which have acquired the capacity to metabolize scFOS have a fitness advantage in the intestine, increasing the risk to human health under FOS pressure.
Supplementary Material
Acknowledgments
We are grateful to Beghin-Meiji (Neuilly/Seine, France) for its generous donation of Profeed P95. We thank Annie Brée and Nathalie Lallier for their excellent assistance in the practical aspects of the work and the staff of the experimental platform for the care of the animals. We thank Roland Quentin and Eric Oswald for providing some strains.
The work was funded by ERA-NET PathoGenoMics (grant ANR-06-PATHO-002-01).
Footnotes
Published ahead of print on 31 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Apanavicius, C. J., K. L. Powell, B. M. Vester, L. K. Karr-Lilienthal, L. L. Pope, N. D. Fastinger, M. A. Wallig, K. A. Tappenden, and K. S. Swanson. 2007. Fructan supplementation and infection affect food intake, fever, and epithelial sloughing from Salmonella challenge in weanling puppies. J. Nutr. 1371923-1930. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, H. J., J.-P. Vaillancourt, and W. B. Gross. 2003. Colibacillosis, p. 631-652. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames.
- 3.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 1008957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, E. L., C. J. Fu, J. H. Porter, and M. S. Kerley. 2005. Fructooligosaccharide supplementation in the yearling horse: effects on fecal pH, microbial content, and volatile fatty acid concentrations. J. Anim. Sci. 831549-1553. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 2771453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bree, A., M. Dho, and J. P. Lafont. 1989. Comparative infectivity for axenic and specific-pathogen-free chickens of O2 Escherichia coli strains with or without virulence factors. Avian Dis. 33134-139. [PubMed] [Google Scholar]
- 7.Buddington, K. K., J. B. Donahoo, and R. K. Buddington. 2002. Dietary oligofructose and inulin protect mice from enteric and systemic pathogens and tumor inducers. J. Nutr. 132472-477. [DOI] [PubMed] [Google Scholar]
- 8.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5376-379. [Google Scholar]
- 9.Chiou, C. Y., H. H. Wang, and G. C. Shaw. 2002. Identification and characterization of the non-PTS fru locus of Bacillus megaterium ATCC 14581. Mol. Genet. Genomics 268240-248. [DOI] [PubMed] [Google Scholar]
- 10.Chouikha, I., P. Germon, A. Brée, P. Gilot, M. Moulin-Schouleur, and C. Schouler. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J. Bacteriol. 188977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collado, M. C., and Y. Sanz. 2007. Characterization of the gastrointestinal mucosa-associated microbiota of pigs and chickens using culture-based and molecular methodologies. J. Food Prot. 702799-2804. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328307-317. [DOI] [PubMed] [Google Scholar]
- 13.Dahlqvist, A., and U. Nilsson. 1984. Cereal fructosans: part 1—isolation and characterization of fructosans from wheat. Food Chem. 14103-112. [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dho, M., and J. P. Lafont. 1982. Escherichia coli colonization of the trachea in poultry: comparison of virulent and avirulent strains in gnotoxenic chickens. Avian Dis. 26787-797. [PubMed] [Google Scholar]
- 16.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30299-316. [PubMed] [Google Scholar]
- 17.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2414-424. [DOI] [PubMed] [Google Scholar]
- 18.Fraenkel, D. G. 1968. The phosphoenolpyruvate-initiated pathway of fructose metabolism in Escherichia coli. J. Biol. Chem. 2436458-6463. [PubMed] [Google Scholar]
- 19.Freter, R., B. Allweiss, P. C. M. O'Brien, S. A. Halstead, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vitro studies. Infect. Immun. 34241-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Germon, P., Y. H. Chen, L. He, J. E. Blanco, A. Bree, C. Schouler, S. H. Huang, and M. Moulin-Schouleur. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 1511179-1186. [DOI] [PubMed] [Google Scholar]
- 21.Goh, Y. J., C. Zhang, A. K. Benson, V. Schlegel, J.-H. Lee, and R. W. Hutkins. 2006. Identification of a putative operon involved in fructooligosaccharide utilization by Lactobacillus paracasei. Appl. Environ. Microbiol. 727518-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong, J., W. Si, R. J. Forster, R. Huang, H. Yu, Y. Yin, C. Yang, and Y. Han. 2007. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 59147-157. [DOI] [PubMed] [Google Scholar]
- 23.González, R., E. S. Klaassens, E. Malinen, W. M. de Vos, and E. E. Vaughan. 2008. Differential transcriptional response of Bifidobacterium longum to human milk, formula milk and galactooligosaccharide. Appl. Environ. Microbiol. 744686-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hacker, J., and E. Carniel. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson, L. A., P. Benson, K. M. Neuzil, M. Grandjean, and J. L. Marino. 2005. Burden of community-onset Escherichia coli bacteremia in seniors. J. Infect. Dis. 1911523-1529. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 662682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khachatryan, Z. A., Z. A. Ktsoyan, G. P. Manukyan, D. Kelly, K. A. Ghazaryan, and R. I. Aminov. 2008. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS ONE 3e3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornberg, H. 2002. If at first you don't succeed—fructose utilization by Escherichia coli. Adv. Enzyme Regul. 42349-360. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence, J. G., and H. Ochman. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA 959413-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Bars, J. 1976. Demonstration of a protocol for obtaining germ-free chickens. Ann. Rech. Vet. 7383-396. (In French.) [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 32.Moulin-Schouleur, M., M. Répérant, S. Laurent, A. Brée, S. Mignon-Grasteau, P. Germon, D. Rasschaert, and C. Schouler. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J. Clin. Microbiol. 453366-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naughton, P. J., L. L. Mikkelsen, and B. B. Jensen. 2001. Effects of nondigestible oligosaccharides on Salmonella enterica serovar Typhimurium and nonpathogenic Escherichia coli in the pig small intestine in vitro. Appl. Environ. Microbiol. 673391-3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parche, S., J. Amon, I. Jankovic, E. Rezzonico, M. Beleut, H. Barutcu, I. Schendel, M. P. Eddy, A. Burkovski, F. Arigoni, and F. Titgemeyer. 2007. Sugar transport systems of Bifidobacterium longum NCC2705. J. Mol. Microbiol. Biotechnol. 129-19. [DOI] [PubMed] [Google Scholar]
- 36.Patterson, J. A., and K. M. Burkholder. 2003. Application of prebiotics and probiotics in poultry production. Poult. Sci. 82627-631. [DOI] [PubMed] [Google Scholar]
- 37.Roberfroid, M. 2007. Prebiotics: the concept revisited. J. Nutr. 137S830-S837. [DOI] [PubMed] [Google Scholar]
- 38.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5449-456. [DOI] [PubMed] [Google Scholar]
- 39.Ryan, S. M., G. F. Fitzgerald, and D. van Sinderen. 2005. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 713475-3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Servin, A. L. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28405-440. [DOI] [PubMed] [Google Scholar]
- 42.Sharp, R., S. Fishbain, and G. T. Macfarlane. 2001. Effect of short-chain carbohydrates on human intestinal bifidobacteria and Escherichia coli in vitro. J. Med. Microbiol. 50152-160. [DOI] [PubMed] [Google Scholar]
- 43.Swanson, K. S., C. M. Grieshop, E. A. Flickinger, L. L. Bauer, J. Chow, B. W. Wolf, K. A. Garleb, and G. C. Fahey, Jr. 2002. Fructooligosaccharides and Lactobacillus acidophilus modify gut microbial populations, total tract nutrient digestibilities and fecal protein catabolite concentrations in healthy adult dogs. J. Nutr. 1323721-3731. [DOI] [PubMed] [Google Scholar]
- 44.Tung, W. L., and K. C. Chow. 1995. A modified medium for efficient electrotransformation of E. coli. Trends Genet. 11128-129. [DOI] [PubMed] [Google Scholar]
- 45.Turk, D. E. 1982. The anatomy of the avian digestive tract as related to feed utilization. Poult. Sci. 611225-1244. [DOI] [PubMed] [Google Scholar]
- 46.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 2992074-2076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.