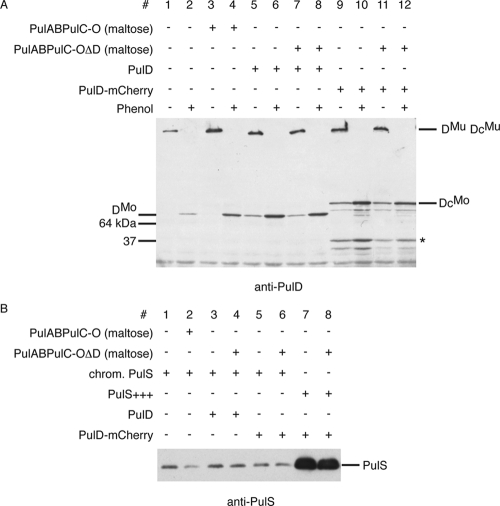
FIG. 4.
Levels of the PulD protein encoded by the maltose-inducible pul operon are comparable to PulD and PulD-mCherry levels obtained by expression of integrated genes at the λatt site and are independent of other Pul factors. (A) Comparison of protein levels of PulD (PAP9124, lanes 5 to 8) and PulD-mCherry (PAP9138, lanes 9 to 12) as multimers (DMu and DcMu, respectively) and as monomers (DMo and DcMo, respectively) (after treatment with phenol) compared to PulD expressed from the pul operon (PAP7232, lanes 1 to 4) with noninduced or induced (maltose) expression of pul genes. The 45-kDa PulD degradation product is indicated by an asterisk. (B) Comparison of chromosomal and plasmid-encoded levels of PulS. The amount of PulS encoded by IPTG-induced pCHAP580 (lanes 7 and 8) is at least 50 times higher than PulS from the constitutively expressed pulS gene in PAP7232 (lanes 1 and 2). Chromosomal levels of PulS are similar in PAP7232 (lanes 1 and 2), PAP9124 (lanes 3 and 4), and PAP9138 (lanes 5 and 6) and are independent of the presence of other Pul factors (maltose).