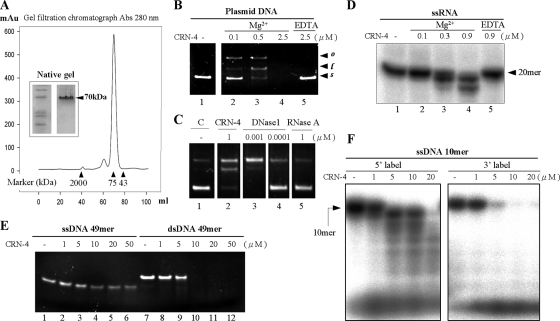
FIG. 2.
In vitro nuclease activity assays of CRN-4. (A) The gel filtration profile of CRN-4 suggested a size of ∼70 kDa, which was about double its calculated molecular mass of 34,641.9 Da, suggesting that CRN-4 was a homodimer in solution in 150 mM NaCl and 50 mM Tris-HCl at pH 7.5. The markers used here include blue dextran (2,000 kDa), conalbumin (75 kDa), and ovalbumin (43 kDa). Native gel analysis of purified CRN-4 also shows that CRN-4 has a molecular mass of ∼70 kDa. (B) CRN-4 digested pQE30 plasmid DNAs in the presence of 1 mM magnesium ions in an enzyme concentration-dependent manner (lanes 2 to 4) but did not digest DNA in the presence of 5 mM EDTA (lane 5). The starting supercoiled substrate (s) was cleaved by CRN-4 to generate linear DNA (l) and open-circle DNA (o). (C) The DNase activity of 1-μM CRN-4 in plasmid digestion is close to that of 0.001 μM DNase I (bovine; Sigma). (D) RNA digestion assays show that a single-stranded 20-mer RNA was digested by CRN-4 in the presence of 1 mM magnesium ions in 100 mM NaCl and 20 mM Tris-HCl (pH 7.5) (lanes 2 to 4). The RNase activity was abolished in the presence of 5 mM EDTA (lane 5). (E) CRN-4 digested a double-stranded 49-mer DNA more efficiently than ssDNA. (F) The ssDNA labeled at either the 5′ or the 3′ end with 32P was incubated with CRN-4. Only the 5′-end-labeled DNA was digested into ladder bands, suggesting that CRN-4 has 3′-5′ exonuclease activity.