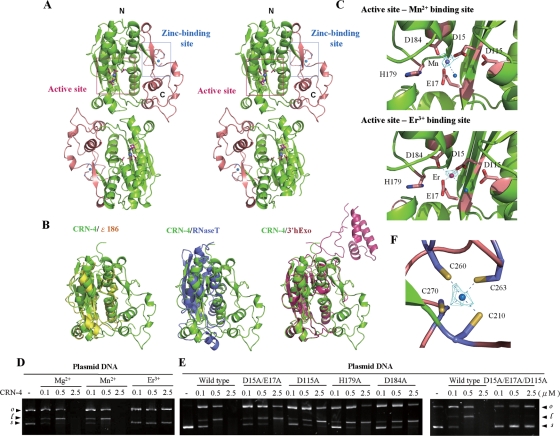
FIG. 4.
Overall structure, structural comparison, and active site of CRN-4. (A) The stereo view of the crystal structure of CRN-4 reveals a homodimeric architecture, with each subunit containing a DEDDh nuclease domain (in green) and a C-terminal Zn-domain (in salmon). The locations of the DEDDh active site and zinc-binding site in one of the subunits are marked by a pink and a blue box, respectively. (B) Superposition of the DEDDh nuclease domain of CRN-4 (green) on those of ɛ186 (yellow, PDB code 1J53), RNase T (blue, PDB code 2IS3), and 3′hExo (red, PDB code 1ZBU). CRN-4 contains an additional Zn-domain compared to ɛ186 and RNase T, while 3′hExo contains an extra SAP domain. (C) The nuclease active sites of Mn-bound and Er-bound CRN-4. The five conserved DEDDh residues in the catalytic domain are displayed in stick models. The blue balls indicate water molecules. The omit 2FoFc maps of Mn2+ and Er3+ are displayed in cyan and contoured at 4.0 σ. (D) CRN-4 was active in the presence of Mg2+ and Mn2+, but inactive with Er3+, as shown by plasmid digestion assays. (E) The CRN-4 mutants D15A/E17A, D115A, H179A, and D184A digested plasmid DNA less efficiently, with activities reduced by ∼5-fold, than did the wild-type CRN-4. The triple mutant D15A/E17A/D115A had almost no observable DNase activity. (F) The zinc ion was bound to four cysteine residues—Cys210, Cys260, Cys263, and Cys270—in the Zn-domain of CRN-4. The omit 2FoFc map of zinc ion is displayed in cyan and contoured at 7.0 σ.