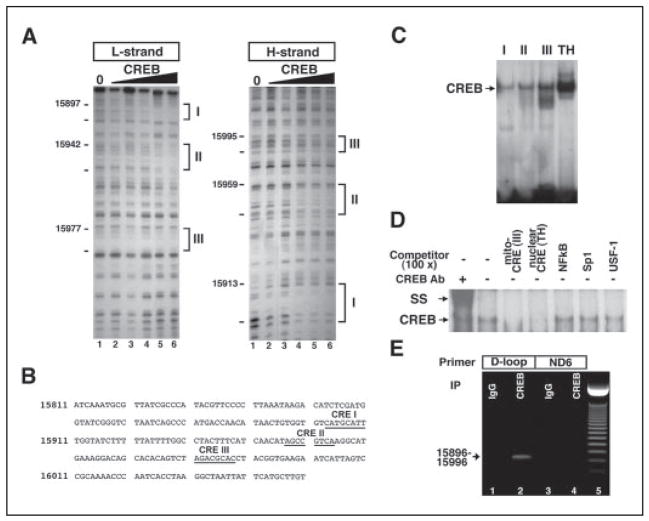
FIGURE 2. CREB binds to the non-coding region of mitochondrial DNA.
A, in vitro footprinting analysis reveals three putative CRE-like sites in the D-loop of mouse mitochondrial DNA. B, sequences of CRE-like sites in the D-loop are presented. Three CRE-like sequences (I–III) as underlined are found in the D-loop. Arrows indicate the direction of CREB binding to the putative CRE. C, EMSA shows that CREB binds to mitochondrial CRE-like sites (I–III). A canonical CRE site in a nuclear gene, tyrosine hydroxylase (TH), promoter was used as a standard of CREB binding activity to mitochondrial DNA. D, nonspecific competitor analysis confirmed that CREB is specifically associated with mitochondrial CRE-like sequences. SS, supershift analysis. E, CREB association with mitochondrial DNA was further examined using DNA-protein cross-linking, immunoprecipitation of pCREB, and PCR methods (see supplemental methods). The mitochondria pellet was cross-linked by 1% formaldehyde, sonicated, and immunoprecipitated. Immuoprecipitated and eluted mitochondrial DNA with CREB antibody and IgG were amplified with primers designed for CRE sites in D-loop sequences and non-CRE sites in ND6 sequences. Lanes 1 and 3, IgG immunoprecipitation; lanes 2 and 4, CREB antibody immunoprecipitation; lane 5, 50-bp molecular marker. The arrow indicates an amplified signal with CRE site primers (15896 –15996).