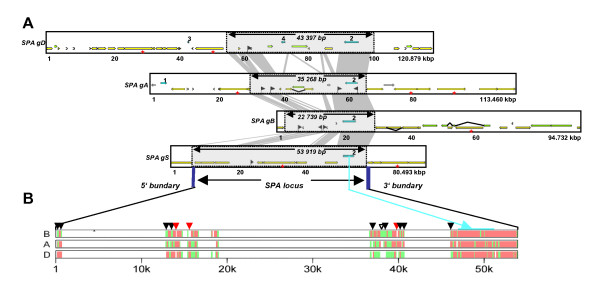
Figure 1.
Identification of the 'SPA orthologous region' and comparative annotation of the homoeologous A, B, D and S sequences. (A) Scaled diagram of annotation results of the SPA locus region in which (CDS) (light blue), class I TEs (yellow blocks), class II TEs (green blocks), unclassified elements (grey), MITEs (vertical black flags) are shown. The remaining white spaces correspond to unassigned DNA (no features of annotation). Grey blocks represent sequence conservation between the different genomes defining the 'SPA orthologous region'. Genes are numbered as follow: 1: Pseudo tubulin gene; 2: SPA; 3: Putative cortical cell-delineating gene; 4: Putative kinesin gene. Eight class I TE displaying complete LTR and TSD suitable for the estimation of the insertion dates are highlighted with red stars. (B) Multipipmaker alignment using the sequence of the SPA orthologous region of the S genome of Ae. speltoides as a matrix compared with the 3 other sequences available, i.e. T. aestivum gB (top), gA (center), gD (bottom). Coloured blocks show the percentage of sequence identity (> 90 in red; between 50 to 90% in green). The SPA gene is indicated as a blue box.