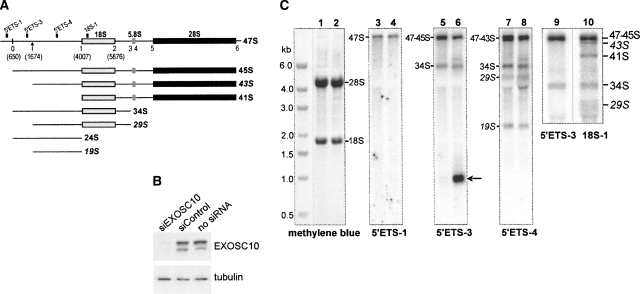
FIGURE 1.
Depletion of EXOSC10 unmasks a second processing site in the 5′ETS of mouse pre-rRNA. (A) Structure of the mouse 47S pre-rRNA transcript showing major cleavage sites and relative position of hybridization probes. Novel pre-rRNAs generated by cleavage at the second site described here (indicated by an arrow) are shown in italics. (B) EXOSC10 protein level is reduced after transfection with specific siRNA, but not after transfections with a heterologous siRNA as compared with mock-transfected cells. Immunoblotting of cell lysates was perfomed with antibodies against PM-Scl100; beta-tubulin was used as a loading control. (C) Hybridization analysis of pre-rRNA. Total RNA was isolated from siControl-transfected (lanes 1,3,5,7,9,10) or siEXOSC10-transfected cells (lanes 2,4,6,8), separated by formaldehyde-agarose gel electrophoresis, blotted on a membrane, stained with methylene blue to reveal 28S, 18S rRNAs, and marker bands, and hybridized with the indicated oligonucleotide probes. Arrow points to the spacer fragment accumulating in EXOSC10-depleted cells. Lanes 9 and 10 show a close-up view of hybridizations in which the gel was run for a longer time to improve separation between 43S and 47/45S pre-rRNAs.