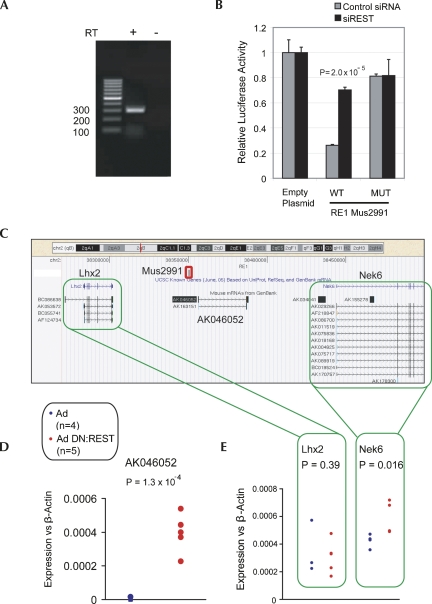
FIGURE 3.
Noncoding transcript AK046052 is targeted by REST. (A) Expression of AK046052 in NS5 was validated by RT-PCR. A control reverse transcription (RT) reaction was carried out without RTase enzyme (−RT). (B) RE1 mus2991 is capable of repressing transcription of a reporter gene in a REST-dependent manner. The RE1 sequence (WT), or a mutated version (MUT), was tested in luciferase assay. The luciferase plasmid, pGL4TK, containing RE1 sequence constructs upstream of the TK promoter was transfected into HEK293 cells in the presence of siRNA targeting REST mRNA (siREST), or control nontargeting siRNA. Luciferase intensity was normalized to that of a control, Renilla plasmid. Ratios are expressed relative to pGL4TK containing no RE1 (Empty). Experiments were carried out on three biological replicates and two technical replicates. Error bars represent the standard error of the mean (SEM), and statistical significance was judged by a Student's t-test. (C) AK046052 resides on mouse Chromosome 2, flanked by known protein-coding genes Lhx2 and Nek6. The validated REST binding site (RE1) mus2991 is located ∼7 kb upstream of AK046052 (red rectangle). (D) The adenovirally delivered dominant-negative REST construct (Ad DN:REST) activated transcription of AK046052 in the neural stem cell line, NS5. Transcript levels of AK046052 were measured using intron-spanning Taqman probes, and normalized to the housekeeping gene, β-actin. Assays were carried out on four control replicates (adenovirus only, Ad) and five dominant-negative replicates (Ad DN:REST). (E) Nek6 was weakly de-repressed by DN:REST, while Lhx2 was unaffected. Expression assays using intron-spanning primers with SYBR green qPCR were carried out on the same samples as in D. Statistical significance was calculated using Student's t-test.