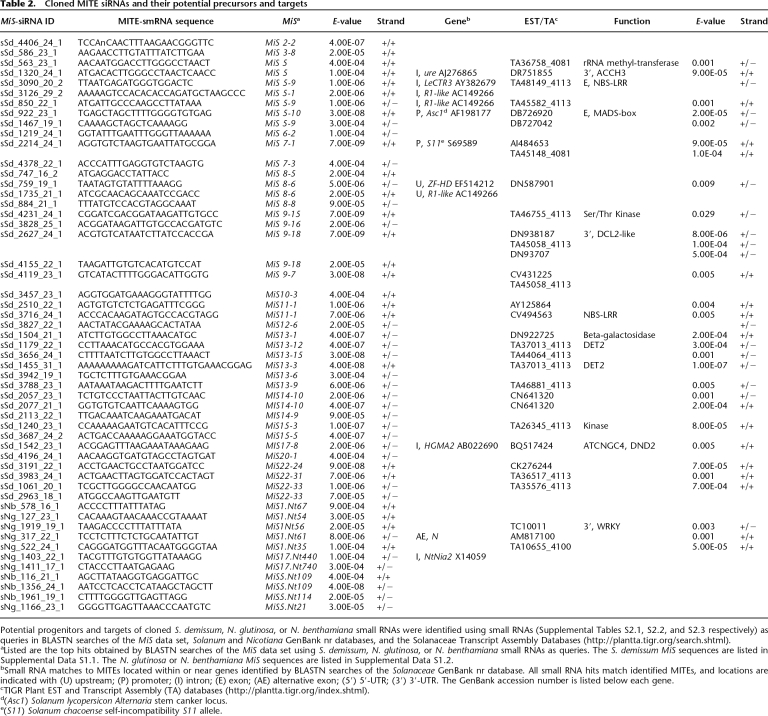
Table 2.
Cloned MITE siRNAs and their potential precursors and targets
Potential progenitors and targets of cloned S. demissum, N. glutinosa, or N. benthamiana small RNAs were identified using small RNAs (Supplemental Tables S2.1, S2.2, and S2.3 respectively) as queries in BLASTN searches of the MiS data set, Solanum and Nicotiana GenBank nr databases, and the Solanaceae Transcript Assembly Databases (http://plantta.tigr.org/search.shtml).
aListed are the top hits obtained by BLASTN searches of the MiS data set using S. demissum, N. glutinosa, or N. benthamiana small RNAs as queries. The S. demissum MiS sequences are listed in Supplemental Data S1.1. The N. glutinosa or N. benthamiana MiS sequences are listed in Supplemental Data S1.2.
bSmall RNA matches to MITEs located within or near genes identified by BLASTN searches of the Solanaceae GenBank nr database. All small RNA hits match identified MITEs, and locations are indicated with (U) upstream; (P) promoter; (I) intron; (E) exon; (AE) alternative exon; (5′) 5′-UTR; (3′) 3′-UTR. The GenBank accession number is listed below each gene.
cTIGR Plant EST and Transcript Assembly (TA) databases (http://plantta.tigr.org/index.shtml).
d(Asc1) Solanum lycopersicon Alternaria stem canker locus.
e(S11) Solanum chacoense self-incompatibility S11 allele.