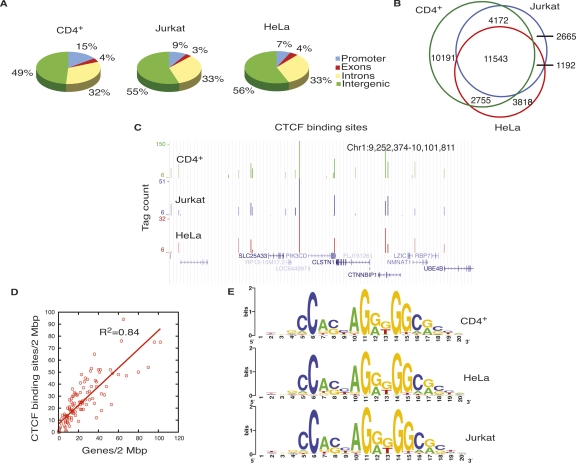
Figure 1.
CTCF-binding sites overlap among different cell types. (A) Genome-wide distribution of CTCF-binding sites in CD4+ T cells, Jurkat cells, and HeLa cells. (B) CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells overlap extensively. (C) Overlap of CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells in an 850-kb region in chromosome 1, shown as custom tracks on the UCSC genome browser (Karolchik et al. 2008). (D) The densities of genes and CTCF-binding sites per 2 Mbp in chromosome 1. X-axis, number of genes/2 Mbp; y-axis, number of CTCF-binding sites/2 Mbp. (E) CTCF-binding motif in CD4+ T cells, HeLa cells, and Jurkat cells.