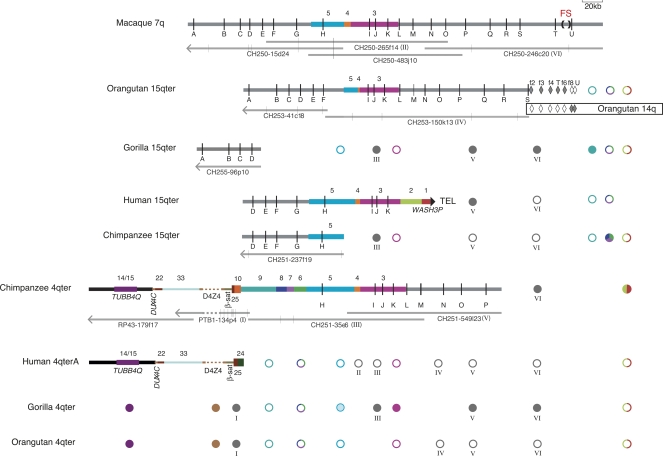
Figure 2.
Sequence content of the fission site region of macaque chromosome 7 and chromosomes 15q and 4q subtelomeres in primates, as compiled from sequenced BACs and targeted FISH and PCR assays. The annotated thick horizontal lines indicate sequence contigs derived from non-human BACs or the human genome assembly. The large filled triangle at the right end of human 15q indicates it as the only one of these sequence contigs to reach the terminal telomere-repeat arrays. Vertical black lines represent OR genes identified by letters A–U (see Supplemental Table 6 for official nomenclature for these genes). The fission site (FS), indicated by (), lies between ORs T and U. Colored bars represent sequence blocks known to be subtelomerically duplicated in the human genome (Linardopoulou et al. 2005); blocks 5, 4, and 3 derive from the fission site region. Horizontal gray bars indicate additional sequences orthologous to the rhesus macaque fission-site region. Orange-brown and gray dashes indicate unsequenced D4Z4 arrays in contigs and BAC PTB1-134p4, respectively. Filled or open diamonds indicate PCR assays on flow-sorted orangutan chromosomes 15 or 14 that were positive or negative, respectively (Supplemental Table 3). The spans of the constituent sequenced BACs are indicated by the thin gray lines under each non-human contig; vertical gray lines on these tracks indicate gaps in sequence contigs; left-pointing arrowheads indicate that the BAC sequence extends toward the centromere from portion shown. BACs used for FISH are indicated by roman numerals I–VI in parentheses. Circles summarize the results of FISH assays for sequence whose presence is not already evident from the sequence contigs; filled or open circles indicate the presence or absence, respectively, of a cumulative signal-intensity score at the cytogenetic location exceeding 25% of the maximum cumulative score observed in the experiment (Trask et al. 1998 for block 3; Supplemental Table 2). Note that the presence of a FISH signal does not necessarily mean that the entire probe sequence resides at that location. Gray circles labeled with roman numerals I–VI represent FISH results with BACs constituting the contigs. Colored circles correspond to block-specific FISH probes; multicolored circles indicate that the FISH probe spanned more than one block. Circles for gorilla 4q represent a compilation of FISH results (Supplemental Table 2) and our PCR assays for blocks 9, 8, 7, 6, and 5 sequences in 4q-derived, D4Z4-containing gorilla BACs identified by Bodega et al. (2007) (Supplemental Table 3). The block-5 circle is partially shaded to indicate that gorilla 4q is FISH negative for this sequence, but these gorilla 4q BACs are PCR positive only for the most distal block 5 assays. Note that FISH assays detect the presence/absence, but not the order, of sequences in a given subtelomere. Wherever possible, FISH-result symbols are aligned with the corresponding probe’s sequence in at least one contig; the FISH symbols for blocks 1/2, 9, and 6/7/8 are shown at the far right to indicate that these probe sequences, when detected by FISH, most likely lie telomeric of the sequence in the contigs. The sequence contigs are drawn to scale and aligned to facilitate comparison of their contents: macaque 7q, orangutan 15q, and human 15q contigs are aligned at OR gene I; chimpanzee 15q and 4q are aligned to human 15q at gene H; gorilla 15q is aligned to human 15q at gene D; human 4q is aligned to chimpanzee 4q at TUBB4Q. The diagrams of sequence contigs are truncated at arrowheads just proximal of gene A to exclude a complex region of tandem repeats that proved difficult to align across species, and the diagram for PTB1-134p4, is truncated at an arrowhead to indicate the extent of curated sequence from this BAC.