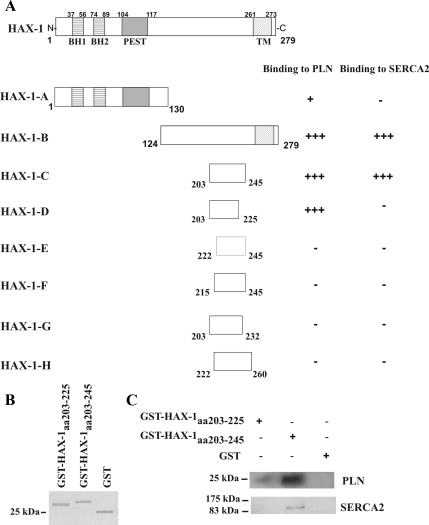
Figure 3.
Determination of HAX-1 minimal binding domain required for PLN or SERCA2 interaction. (A) To delineate the HAX-1 C-terminal region required for binding to either PLN or SERCA2, a series of HAX-1 deletion constructs were generated and tested for their ability to bind to PLN or SERCA2 in yeast. Amino acids 203-225 are required for binding to PLN, whereas a larger region of HAX-1, encompassing amino acids 203-245, is required for its binding to SERCA2. The Bcl-2 homology domains 1 and 2 (BH1 and BH2), PEST sequence and transmembrane domain (TM) of HAX-1 are indicated. (B) Recombinant GST-HAX-1aa 203-225 (∼28 kDa), GST-HAX-1aa 203-245 (∼30 kDa) and GST (∼25 kDa) proteins were subjected to SDS-PAGE and stained with Coomassie Blue. (C) To confirm the findings from the yeast two-hybrid system, GST pull-down assays were performed using equivalent amounts of GST-HAx-1aa 203-225, GST-HAX-1aa 203-245, and GST-protein bound to glutathione matrices and cardiac homogenates. Immunoblot analysis using anti-PLN or anti-SERCA2 antibodies determined that GST-HAX-1aa 203-245 binds to both PLN and SERCA2 native proteins, whereas GST- HAx-1aa 203-225 binds only to PLN.