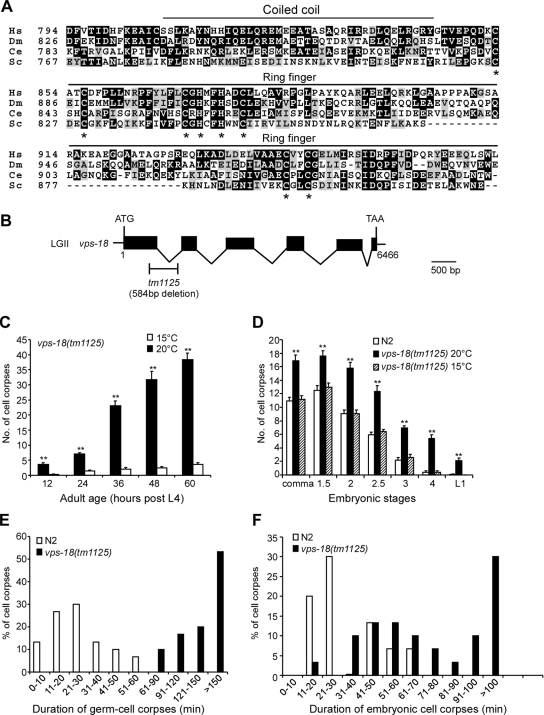
Figure 1.
C. elegans vps-18 affects cell corpse clearance. (A) Sequence alignment of the C-terminal region of VPS proteins of human (Hs), Drosophila (Dm), C. elegans (Ce), and yeast (Sc). The identical amino acids are shaded in black and similar amino acids are shaded in gray. The top lines indicate the regions for coiled-coil domain and RING-finger motif. Asterisks indicate the residues critical for RING-finger motif. (B) Schematic representation of vps-18(tm1125) deletion. Solid boxes indicate exons and waved lines indicate introns. The fragment below the gene indicates the region and size of the deletion mutation of vps-18. The scale bar represents 500 base pairs. (C) The profile of germ cell death in vps-18(tm1125) mutants. Germ cell corpses in one gonad arm of each animal were scored every 12 h after L4 stage. The error bars represent SEM. Fifteen animals were scored for each time point. Comparisons were performed with unpaired t test. Single asterisks indicate p < 0.05 and double asterisks indicate p < 0.001. (D) The profile of embryonic cell death in vps-18(tm1125) mutants. Cell corpses of 15 embryos at each embryonic stage were scored. Comparisons were performed as described in C. (E and F) Four-dimensional microscopy analysis of durations of persistence of germ cell corpses (E) and embryonic cell corpses (F) in N2 and vps-18(tm1125) animals. The duration of 30 germ cell corpses and 30 embryonic cell corpses in N2 and vps-18(tm1125) animals were recorded, respectively.