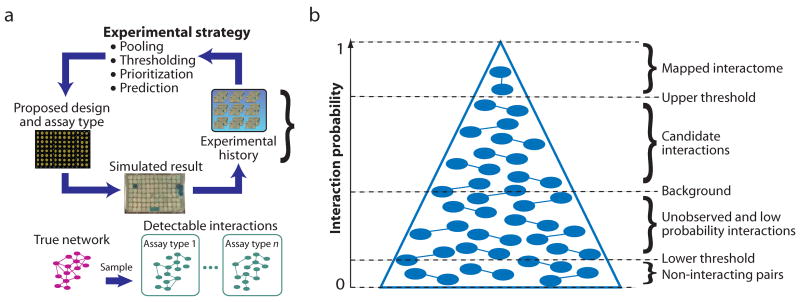
Figure 1. Simulating an interaction mapping project.
(a) At any given point in the project, every pair of proteins is assigned an interaction probability based on its experimental history (initially these probabilities are set to background or informed by predictions). The interaction probabilities and experimental history are used to design a 96-well plate Y2H experiment according to one of the strategies. The result of this experiment is simulated based on the detectability of the tested interactions (given the assay type) and the pooling sensitivity. The new experimental results are recorded in the history and also (b) used to update the interaction probabilities of the relevant protein pairs. The pyramid represents the ordered list of protein pairs ranked by probability. It is wider at the bottom than at the top to reflect that most pairs are negative—i.e., most pairs will have low probability and only a few pairs will percolate to the top of the list with high probability. Interactions with probability above an upper threshold are added to the mapped interactome, which is compared to the simulated “True Network” at intervals of 1,000 plates for reporting coverage and FDR.